Fast and performant TCR representation model
Project description
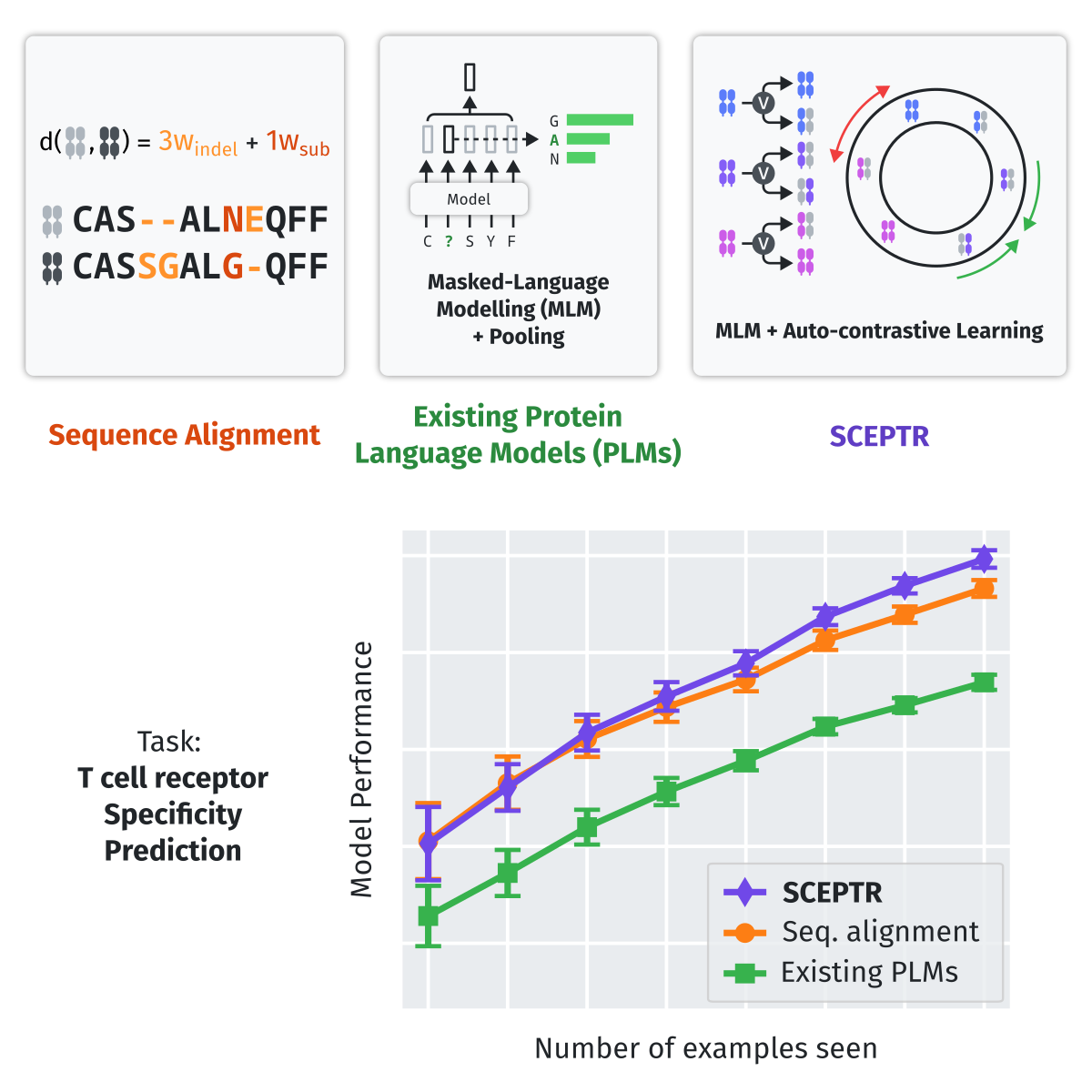 |
|---|
| Graphical abstract. Traditional protein language models that are trained purely on masked-language modelling underperform sequence alignment models on TCR specificity prediction. In contrast, our model SCEPTR is jointly trained on masked-language modelling and contrastive learning, allowing it to outperform other language models as well as the best sequence alignment models to achieve state-of-the-art performance. |
SCEPTR (Simple Contrastive Embedding of the Primary sequence of T cell Receptors) is a small, fast, and informative TCR representation model that can be used for alignment-free TCR analysis, including for TCR-pMHC interaction prediction and TCR clustering (metaclonotype discovery). Our manuscript demonstrates that SCEPTR can be used for few-shot TCR specificity prediction with improved accuracy over previous methods.
SCEPTR is a BERT-like transformer-based neural network implemented in Pytorch. With the default model providing best-in-class performance with only 153,108 parameters (typical protein language models have tens or hundreds of millions), SCEPTR runs fast- even on a CPU! And if your computer does have a CUDA-enabled GPU, the sceptr package will automatically detect and use it, giving you blazingly fast performance without the hassle.
sceptr's API exposes four intuitive functions: calc_cdist_matrix, calc_pdist_vector, calc_vector_representations, and calc_residue_representations -- and it's all you need to make full use of the SCEPTR models.
What's even better is that they are fully compliant with pyrepseq's tcr_metric API, so sceptr will fit snugly into the rest of your repertoire analysis workflow.
Installation
pip install sceptr
Citing SCEPTR
Please cite our manuscript.
BibTex
@article{nagano_contrastive_2025,
title = {Contrastive learning of {T} cell receptor representations},
volume = {16},
issn = {2405-4712, 2405-4720},
url = {https://www.cell.com/cell-systems/abstract/S2405-4712(24)00369-7},
doi = {10.1016/j.cels.2024.12.006},
language = {English},
number = {1},
urldate = {2025-01-19},
journal = {Cell Systems},
author = {Nagano, Yuta and Pyo, Andrew G. T. and Milighetti, Martina and Henderson, James and Shawe-Taylor, John and Chain, Benny and Tiffeau-Mayer, Andreas},
month = jan,
year = {2025},
pmid = {39778580},
note = {Publisher: Elsevier},
keywords = {contrastive learning, protein language models, representation learning, T cell receptor, T cell specificity, TCR, TCR repertoire},
}
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
Filter files by name, interpreter, ABI, and platform.
If you're not sure about the file name format, learn more about wheel file names.
Copy a direct link to the current filters
File details
Details for the file sceptr-1.2.0.tar.gz.
File metadata
- Download URL: sceptr-1.2.0.tar.gz
- Upload date:
- Size: 11.1 MB
- Tags: Source
- Uploaded using Trusted Publishing? Yes
- Uploaded via: twine/6.1.0 CPython/3.13.7
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
0f9e0b5630e0d032c95ecb316d6431374ef51e83be3d16b190139c87e3df5551
|
|
| MD5 |
dcbc6e257d657e35c966e3f84be0d937
|
|
| BLAKE2b-256 |
bda42f534e3cab4a0336c7d8646140e4ba2cdc4717cd9cb5f76aa5cd77d9e708
|
Provenance
The following attestation bundles were made for sceptr-1.2.0.tar.gz:
Publisher:
publish_to_pypi.yaml on yutanagano/sceptr
-
Statement:
-
Statement type:
https://in-toto.io/Statement/v1 -
Predicate type:
https://docs.pypi.org/attestations/publish/v1 -
Subject name:
sceptr-1.2.0.tar.gz -
Subject digest:
0f9e0b5630e0d032c95ecb316d6431374ef51e83be3d16b190139c87e3df5551 - Sigstore transparency entry: 698549707
- Sigstore integration time:
-
Permalink:
yutanagano/sceptr@ea7f68cdce74c15bc8a2f776a4688f58eb9b2eb7 -
Branch / Tag:
refs/tags/v1.2.0 - Owner: https://github.com/yutanagano
-
Access:
public
-
Token Issuer:
https://token.actions.githubusercontent.com -
Runner Environment:
github-hosted -
Publication workflow:
publish_to_pypi.yaml@ea7f68cdce74c15bc8a2f776a4688f58eb9b2eb7 -
Trigger Event:
release
-
Statement type:
File details
Details for the file sceptr-1.2.0-py3-none-any.whl.
File metadata
- Download URL: sceptr-1.2.0-py3-none-any.whl
- Upload date:
- Size: 10.9 MB
- Tags: Python 3
- Uploaded using Trusted Publishing? Yes
- Uploaded via: twine/6.1.0 CPython/3.13.7
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
aba652a9fb53861d5170cc1ad99a284852da8899328f4b4fd80e78f595d250d5
|
|
| MD5 |
8ac19301bc55d972e00cf233d8ad3af7
|
|
| BLAKE2b-256 |
8d8ffbca69c18292a8ce8fa1ee4bf9b2ccaf6b10bf93b025a0307d6c669c0355
|
Provenance
The following attestation bundles were made for sceptr-1.2.0-py3-none-any.whl:
Publisher:
publish_to_pypi.yaml on yutanagano/sceptr
-
Statement:
-
Statement type:
https://in-toto.io/Statement/v1 -
Predicate type:
https://docs.pypi.org/attestations/publish/v1 -
Subject name:
sceptr-1.2.0-py3-none-any.whl -
Subject digest:
aba652a9fb53861d5170cc1ad99a284852da8899328f4b4fd80e78f595d250d5 - Sigstore transparency entry: 698549716
- Sigstore integration time:
-
Permalink:
yutanagano/sceptr@ea7f68cdce74c15bc8a2f776a4688f58eb9b2eb7 -
Branch / Tag:
refs/tags/v1.2.0 - Owner: https://github.com/yutanagano
-
Access:
public
-
Token Issuer:
https://token.actions.githubusercontent.com -
Runner Environment:
github-hosted -
Publication workflow:
publish_to_pypi.yaml@ea7f68cdce74c15bc8a2f776a4688f58eb9b2eb7 -
Trigger Event:
release
-
Statement type:

















