Implementation of the Empirical Mode Decomposition (EMD) and its variations
Project description
PyEMD
Links
- HTML documentation: https://pyemd.readthedocs.org
- Issue tracker: https://github.com/laszukdawid/pyemd/issues
- Source code repository: https://github.com/laszukdawid/pyemd
Introduction
This is yet another Python implementation of Empirical Mode Decomposition (EMD). The package contains many EMD variations and intends to deliver more in time.
EMD variations:
- Ensemble EMD (EEMD),
- "Complete Ensemble EMD" (CEEMDAN)
- different settings and configurations of vanilla EMD.
- Image decomposition (EMD2D & BEMD) (experimental, no support)
PyEMD allows to use different splines for envelopes, stopping criteria and extrema interpolation.
Available splines:
- Natural cubic [default]
- Pointwise cubic
- Akima
- Linear
Available stopping criteria:
- Cauchy convergence [default]
- Fixed number of iterations
- Number of consecutive proto-imfs
Extrema detection:
- Discrete extrema [default]
- Parabolic interpolation
Installation
Recommended
Simply download this directory either directly from GitHub, or using command line:
$ git clone https://github.com/laszukdawid/PyEMD
Then go into the downloaded project and run from command line:
$ python setup.py install
PyPi
Packaged obtained from PyPi is/will be slightly behind this project, so some features might not be the same. However, it seems to be the easiest/nicest way of installing any Python packages, so why not this one?
$ pip install EMD-signal
Example
More detailed examples are included in the documentation or in the PyEMD/examples.
EMD
In most cases default settings are enough. Simply import EMD and pass
your signal to instance or to emd() method.
from PyEMD import EMD
import numpy as np
s = np.random.random(100)
emd = EMD()
IMFs = emd(s)
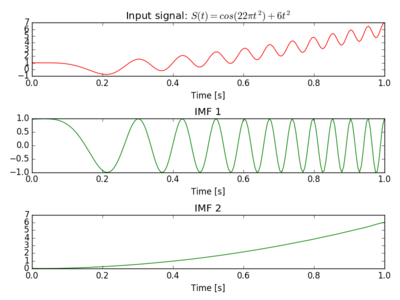
The Figure below was produced with input: $S(t) = cos(22 \pi t^2) + 6t^2$
EEMD
Simplest case of using Ensemble EMD (EEMD) is by importing EEMD and
passing your signal to the instance or eemd() method.
Windows: Please don't skip the if __name__ == "__main__" section.
from PyEMD import EEMD
import numpy as np
if __name__ == "__main__":
s = np.random.random(100)
eemd = EEMD()
eIMFs = eemd(s)
CEEMDAN
As with previous methods, there is also simple way to use CEEMDAN.
Windows: Please don't skip the if __name__ == "__main__" section.
from PyEMD import CEEMDAN
import numpy as np
if __name__ == "__main__":
s = np.random.random(100)
ceemdan = CEEMDAN()
cIMFs = ceemdan(s)
Visualisation
The package contain a simple visualisation helper that can help, e.g., with time series and instantaneous frequencies.
import numpy as np
from PyEMD import EMD, Visualisation
t = np.arange(0, 3, 0.01)
S = np.sin(13*t + 0.2*t**1.4) - np.cos(3*t)
# Extract imfs and residue
# In case of EMD
emd = EMD()
emd.emd(S)
imfs, res = emd.get_imfs_and_residue()
# In general:
#components = EEMD()(S)
#imfs, res = components[:-1], components[-1]
vis = Visualisation()
vis.plot_imfs(imfs=imfs, residue=res, t=t, include_residue=True)
vis.plot_instant_freq(t, imfs=imfs)
vis.show()
EMD2D/BEMD
Unfortunately, this is Experimental and we can't guarantee that the output is meaningful.
The simplest use is to pass image as monochromatic numpy 2D array. Sample as
with the other modules one can use the default setting of an instance or, more explicitly,
use the emd2d() method.
from PyEMD.EMD2d import EMD2D #, BEMD
import numpy as np
x, y = np.arange(128), np.arange(128).reshape((-1,1))
img = np.sin(0.1*x)*np.cos(0.2*y)
emd2d = EMD2D() # BEMD() also works
IMFs_2D = emd2d(img)
F.A.Q
Why is EEMD/CEEMDAN so slow?
Unfortunately, that's their nature. They execute EMD multiple times every time with slightly modified version. Added noise can cause a creation of many extrema which will decrease performance of the natural cubic spline. For some tweaks on how to deal with that please see Speedup tricks in the documentation.
Contact
Feel free to contact me with any questions, requests or simply to say hi. It's always nice to know that I've helped someone or made their work easier. Contributing to the project is also acceptable and warmly welcomed.
Contact me either through gmail (laszukdawid @ gmail) or search me through your favourite web search.
Citation
If you found this package useful and would like to cite it in your work please use the following structure:
@misc{pyemd,
author = {Laszuk, Dawid},
title = {Python implementation of Empirical Mode Decomposition algorithm},
year = {2017},
publisher = {GitHub},
journal = {GitHub Repository},
howpublished = {\url{https://github.com/laszukdawid/PyEMD}},
}
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
File details
Details for the file EMD-signal-0.2.15.tar.gz.
File metadata
- Download URL: EMD-signal-0.2.15.tar.gz
- Upload date:
- Size: 49.7 kB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/3.3.0 pkginfo/1.7.0 requests/2.24.0 setuptools/52.0.0 requests-toolbelt/0.9.1 tqdm/4.56.0 CPython/3.8.5
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 263f3719f7b2ff656a9e7e651059dc1e1d22ffaabd37134f1477af1cdf29d7bf |
|
| MD5 | 2b3fc730ac784ff335ea1c4f85ee8aac |
|
| BLAKE2b-256 | 75a156a66120332097353491234c5a9e9ad71173d012137369bda46ea7181a3e |
File details
Details for the file EMD_signal-0.2.15-py2.py3-none-any.whl.
File metadata
- Download URL: EMD_signal-0.2.15-py2.py3-none-any.whl
- Upload date:
- Size: 42.1 kB
- Tags: Python 2, Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/3.3.0 pkginfo/1.7.0 requests/2.24.0 setuptools/52.0.0 requests-toolbelt/0.9.1 tqdm/4.56.0 CPython/3.8.5
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 31540d1c59a7e91b98b284c23cf0e6ce54fa01a0e6a0f41b4d937283b3c4a42a |
|
| MD5 | 54b06d01ae7faf7b3ae2c62eb27c0641 |
|
| BLAKE2b-256 | 4d4137856c480e5fb1d4fc151879557bb4b38f87378d8879b4f0794d1999f5c5 |