SpatialDM: Spatial co-expression Detected by bivariate Moran
Project description
About
SpatialDM (Spatial Direct Messaging, or Spatial co-expressed ligand and receptor Detected by Moran’s bivariant extension), a statistical model and toolbox to identify the spatial co-expression (i.e., spatial association) between a pair of ligand and receptor.
Uniquely, SpatialDM can distinguish co-expressed ligand and receptor pairs from spatially separating pairs, and identify the spots of interaction.
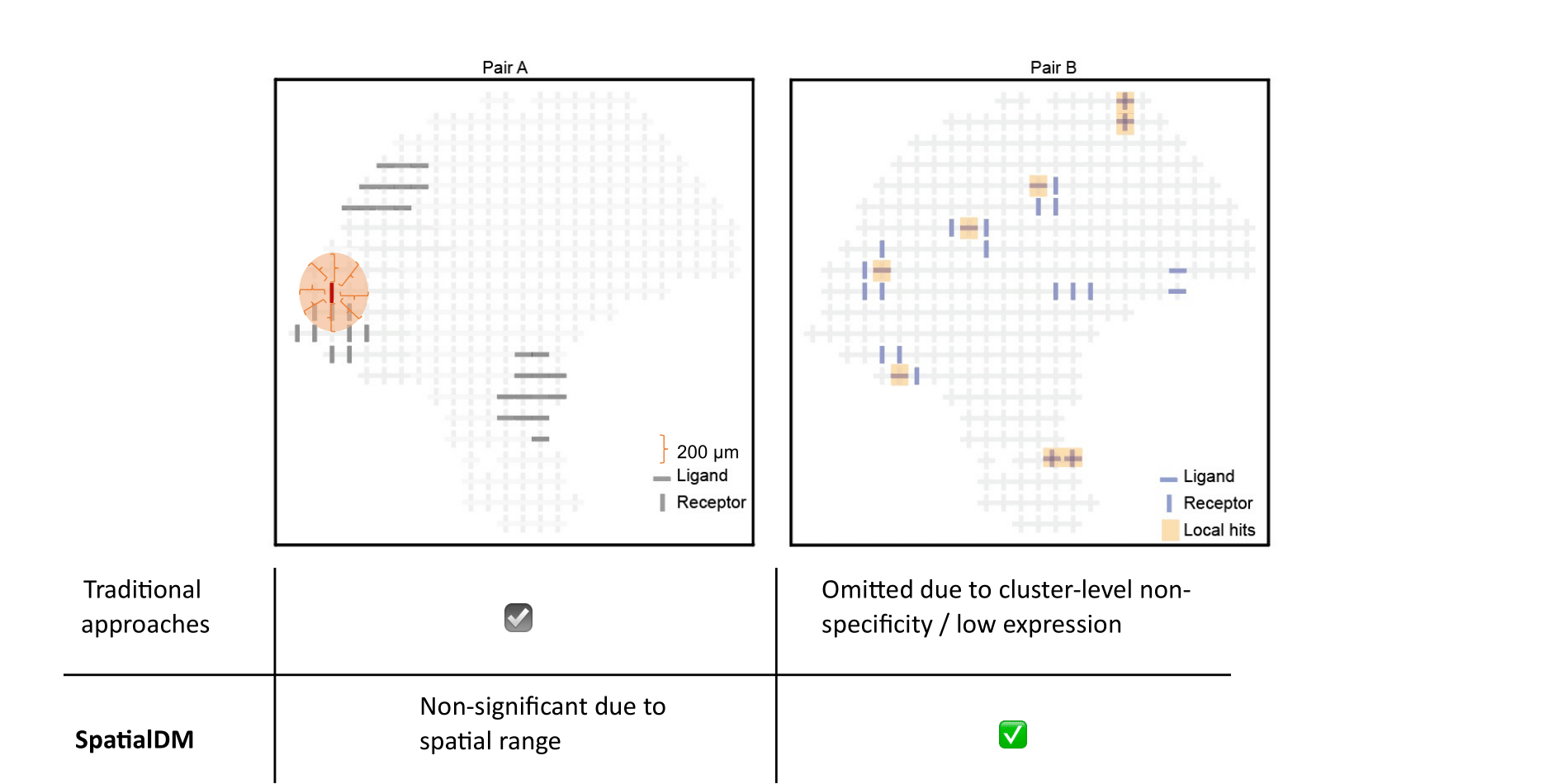
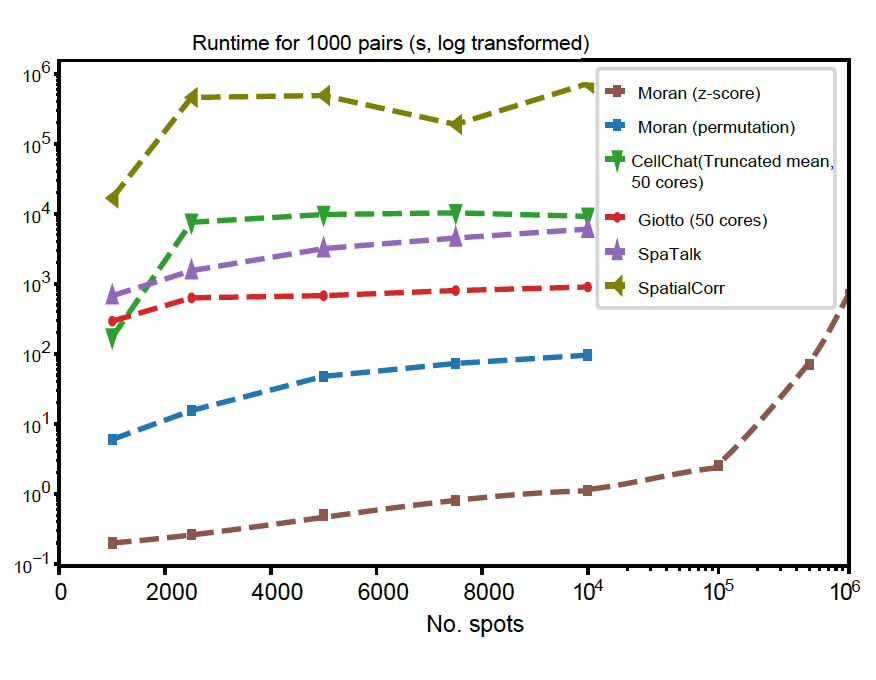
With the analytical testing method, SpatialDM is scalable to 1 million spots within 12 min with only one core.
It comprises two main steps: 1) global selection spatialdm_global to identify significantly interacting LR pairs; 2) local selection spatialdm_local to identify local spots for each interaction.
Installation
SpatialDM is available through PyPI. To install, type the following command line and add -U for updates:
pip install -U SpatialDMAlternatively, you can install from this GitHub repository for latest (often development) version by the following command line:
pip install -U git+https://github.com/StatBiomed/SpatialDMInstallation time: < 1 min
Quick example
Using the build-in melanoma dataset as an example, the following Python script will compute the p-value indicating whether a certain Ligand-Receptor is spatially co-expressed.
import spatialdm as sdm
adata = sdm.datasets.dataset.melanoma()
sdm.weight_matrix(adata, l=1.2, cutoff=0.2, single_cell=False) # weight_matrix by rbf kernel
sdm.extract_lr(adata, 'human', min_cell=3) # find overlapping LRs from CellChatDB
sdm.spatialdm_global(adata, 1000, specified_ind=None, method='both', nproc=1) # global Moran selection
sdm.sig_pairs(adata, method='permutation', fdr=True, threshold=0.1) # select significant pairs
sdm.spatialdm_local(adata, n_perm=1000, method='both', specified_ind=None, nproc=1) # local spot selection
sdm.sig_spots(adata, method='permutation', fdr=False, threshold=0.1) # significant local spots
# visualize global and local pairs
import spatialdm.plottings as pl
pl.global_plot(adata, pairs=['SPP1_CD44'])
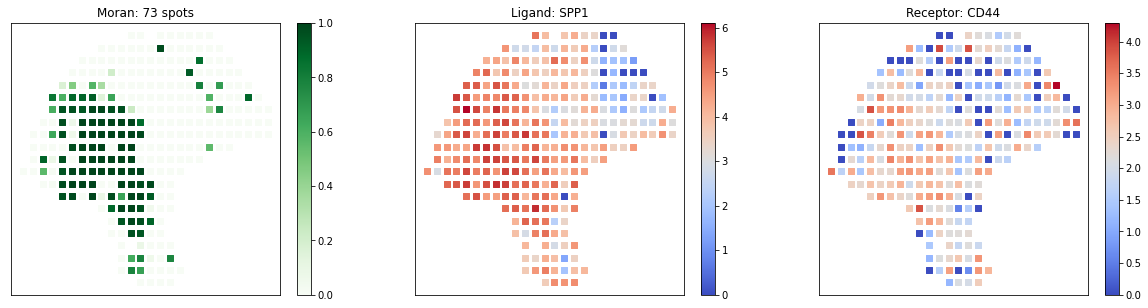
pl.plot_pairs(adata, ['SPP1_CD44'], marker='s')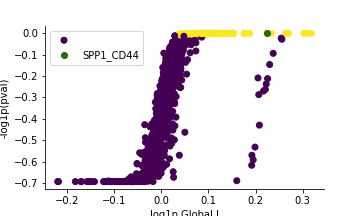
Detailed Manual
The full manual is at https://spatialdm.readthedocs.io, including:
References
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
File details
Details for the file spatialdm-0.3.1.tar.gz.
File metadata
- Download URL: spatialdm-0.3.1.tar.gz
- Upload date:
- Size: 5.7 MB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/5.0.0 CPython/3.11.3
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
2445eeec8fca557cf9b86d0af03a6658287796a37dc3e7e9117461176cc0892f
|
|
| MD5 |
6efff2433a678b5319a3fd21a833ccc4
|
|
| BLAKE2b-256 |
9f10792b87e5f9a6cbf8bbc04486231a54ca200b48744bd1cfaa2af531af38c9
|












