script to generate amplicon coverage plot
Project description
amplicon_coverage_plot
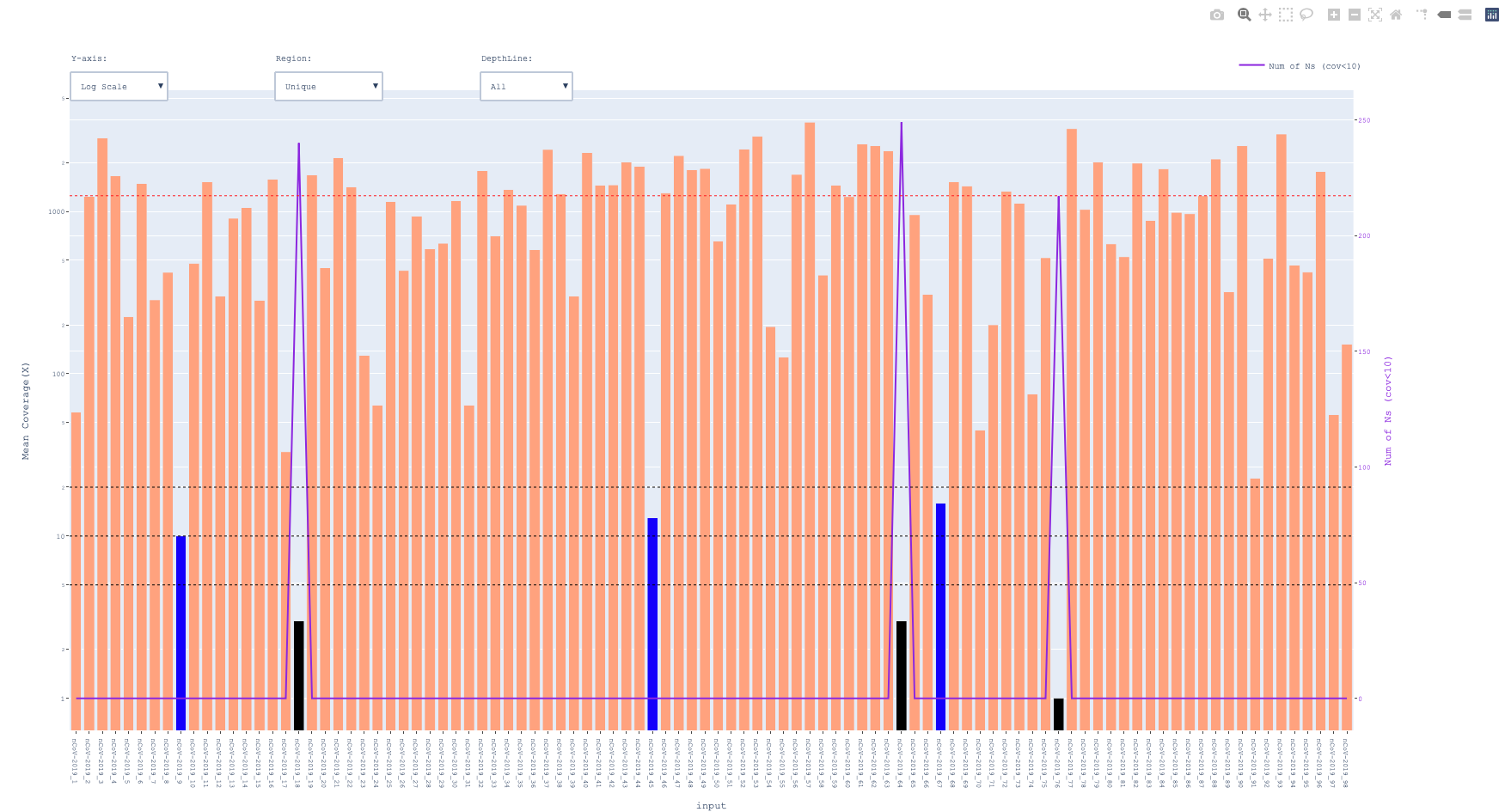
The script will generate an interactive barplot given amplicon info in bed6/bedpe format and coverage information in cov/bam file.
Dependencies
Programming/Scripting languages
- Python >=v3.7
- The pipeline has been tested in v3.7.6
Python packages
Third party softwares/packages
- samtools >=1.9 - process bam file
Installation
Install by pip
pip install amplicov
Install by conda
conda install -c bioconda amplicon_coverage_plot
Install from source
Clone the amplicon_coverage_plot repository.
git clone https://github.com/chienchi/amplicon_coverage_plot
Then change directory to amplicon_coverage_plot and install.
cd amplicon_coverage_plot
python setup.py install
If the installation was succesful, you should be able to type amplicov -h and get a help message on how to use the tool.
amplicov -h
Usage
usage: amplicov [-h] (--bed [FILE] | --bedpe [FILE])
(--bam [FILE] | --cov [FILE]) [-o [PATH]] [-p [STR]] [--pp]
[--mincov [INT]] [--version]
Script to parse amplicon region coverage and generate barplot in html
optional arguments:
-h, --help show this help message and exit
--pp process proper paired only reads from bam file
(illumina)
--mincov [INT] minimum coverage to count as ambiguous N site
[default:10]
-r [STR], --refID [STR]
reference accession (bed file first field)
--depth_lines DEPTH_LINES [DEPTH_LINES ...]
Add option to display lines at these depths (provide depths as a list of integers) [default:5 10 20 50]
--gff [FILE] gff file for data hover info annotation
--version show program's version number and exit
Amplicon Input (required, mutually exclusive):
--bed [FILE] amplicon bed file (bed6 format)
--bedpe [FILE] amplicon bedpe file
Coverage Input (required, mutually exclusive):
--bam [FILE] sorted bam file (ex: samtools sort input.bam -o
sorted.bam)
--cov [FILE] coverage file [position coverage]
Output:
-o [PATH], --outdir [PATH]
output directory
-p [STR], --prefix [STR]
output prefix
Test
cd tests
./runTest.sh
Outputs
-- prefix_amplicon_coverage.txt
| ID | Whole_Amplicon | Unique | Whole_Amplicon_Ns(cov<10) | Unique_Amplicon_Ns(cov<10) |
|---|---|---|---|---|
| nCoV-2019_1 | 217.74 | 53.00 | 0.00 | 0.00 |
| nCoV-2019_2 | 1552.83 | 1235.50 | 0.00 | 0.00 |
| nCoV-2019_3 | 3164.22 | 2831.73 | 0.00 | 0.00 |
| nCoV-2019_4 | 2005.16 | 1658.00 | 0.00 | 0.00 |
| etc... |
Table Header Definition in the amplicon_coverage.txt
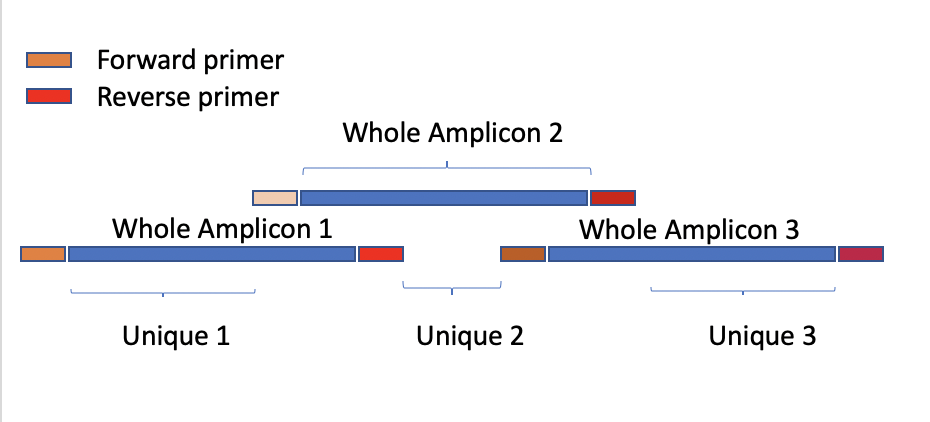
-
Whole_Amplicon_Ns(cov<10): The number of aligned position with coverage < 10 or (--mincov) in the Whole Amplicon region
-
Unique_Amplicon_Ns(cov<10): The number of aligned position with coverage < 10 or (--mincov) in the Unique region
-- prefix_amplicon_coverage.html
color black for < 5x and blue for <20x
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.