No project description provided
Project description
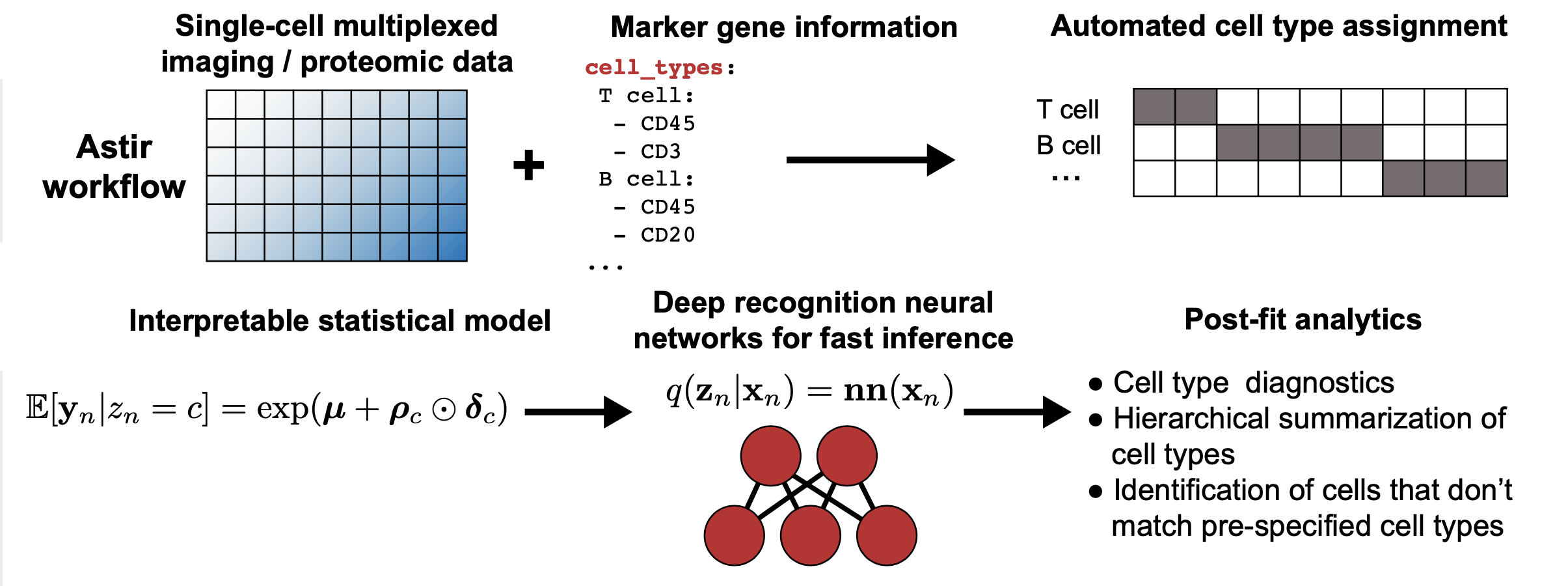
astir is a modelling framework for the assignment of cell type across a range of single-cell technologies such as Imaging Mass Cytometry (IMC). astir is built using pytorch and uses recognition networks for fast minibatch stochastic variational inference.
Key applications:
Automated assignment of cell type and state from highly multiplexed imaging and proteomic data
Diagnostic measures to check quality of resulting type and state inferences
Ability to map new data to cell types and states trained on existing data using recognition neural networks
A range of plotting and data loading utilities
Getting started
Launch the interactive tutorial: 
See the full documentation and check out the tutorials.
Project details
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
Filter files by name, interpreter, ABI, and platform.
If you're not sure about the file name format, learn more about wheel file names.
Copy a direct link to the current filters
File details
Details for the file astir-0.1.5.tar.gz.
File metadata
- Download URL: astir-0.1.5.tar.gz
- Upload date:
- Size: 41.5 kB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/6.1.0 CPython/3.12.2
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
373521b92e715a7e5945925099b8af75daf6686a1d693817a364687a3938d177
|
|
| MD5 |
603033f0126457e59f21bd444d3b98f3
|
|
| BLAKE2b-256 |
001131ffef71ce37df0d8b077e060ed5ccaa5ac9129b1c4a37a9c7c400cce0dc
|
File details
Details for the file astir-0.1.5-py3-none-any.whl.
File metadata
- Download URL: astir-0.1.5-py3-none-any.whl
- Upload date:
- Size: 49.0 kB
- Tags: Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/6.1.0 CPython/3.12.2
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
436e544e67167fb9139195976afa0ea81bf1aac63a28eeeda2f96f0287810c91
|
|
| MD5 |
b1462cd5ce63f24a4e2d66d6e86e5809
|
|
| BLAKE2b-256 |
45e59e8739d3d7e99cf14a43c7ed9ed5f6ca70aa61fec110d992111639491673
|















