Automated version of Extended Aperture Photometry developed for K2 RR Lyrae stars.
Project description
autoEAP - Automated Extended Aperture Photometry, developed for high amplitude K2 variable stars
The details of Extended Aperture Photometry are published in Plachy et al.,2019,ApJS,244,32. A short summary of automatization is published here.
Installation
To install the package, use:
pip install autoeap
Example usage
To create your own photomery, you'll need a Target Pixel File, such as this one. Then, after starting Python, you can do:
yourtpf = '/path/to/your/tpf/ktwo212466080-c17_lpd-targ.fits'
import autoeap
time, flux, flux_err = autoeap.createlightcurve(yourtpf)
Or if you want to let autoEAP download the TPF from MAST database, you can just provide a target name and a campaign number:
import autoeap
targetID = 'EPIC 212466080'
campaign = 17
time, flux, flux_err = autoeap.createlightcurve(targetID,campaign=campaign)
With this last line, you can create autoEAP photometry for any K2 variable star.
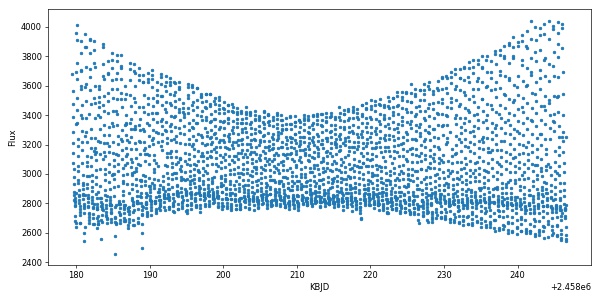
Plotting our results gives:
import matplotlib.pyplot as plt
plt.figure(figsize=(10,5))
plt.plot(time,flux,'.')
plt.xlabel('KBJD')
plt.ylabel('Flux')
plt.show()
The details of the workflow is described in docs.
You can find Google Colab friendly tutorials in the examples.
Apply K2 Systematics Correction (K2SC)
If you want to apply K2SC correction for your freshly made raw-photometry, first you should install K2SC. AutoEAP is based on that package, so if you find K2SC useful, please cite Aigrain et al.,2016,MNRAS,459,2408.
Installation:
git clone https://github.com/OxES/k2sc.git
cd k2sc
python setup.py install --user
If it fails, try to install it with george:
pip3 install george k2sc
And then without much hassle, you can use in python:
import autoeap
time, flux, flux_err = autoeap.createlightcurve(yourtpf,apply_K2SC=True)
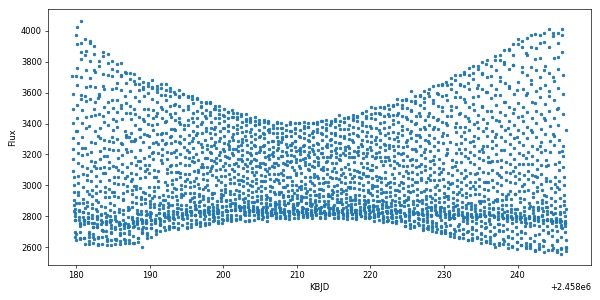
The result is quite delightful:
import matplotlib.pyplot as plt
plt.figure(figsize=(10,5))
plt.plot(time,flux,'.')
plt.xlabel('KBJD')
plt.ylabel('Flux')
plt.show()
Apply spline correction
We have also built-in a method to remove trends using low-order splines. Just do to correct the raw light curve:
import autoeap
time, flux, flux_err = autoeap.createlightcurve(yourtpf,remove_spline=True)
Or do this to remove a spline from the K2SC light curve:
import autoeap
time, flux, flux_err = autoeap.createlightcurve(yourtpf,apply_K2SC=True,remove_spline=True)
Available options
apply_K2SCIfTrue, after the raw photomery, K2SC will be applied to remove systematics from the extracted light curve.remove_splineIfTrue, after the raw photomery, a low-order spline will be fitted and removed from the extracted light curve. Ifapply_K2SCis alsoTrue, then this step will be done after the K2SC.save_lcIfTrue, the intermediate and the final light curves will be save as a file.campaignIf local TPF file is not found, it will be downloaded from MAST, butcampaignnumber should be defined as well, if the target has been observed in more than one campaign. Otherwise that campaign will be used in which the target were first observed.show_plotsIfTrue, all the plots will be displayed.save_plotsIfTrue, all the plots will be saved to a subdirectory.window_lengthThe length of filter window for spline correction given in days. Applies only ifremove_splineisTrue. Default is20days.sigma_lowerThe number of standard deviations to use as the lower bound for sigma clipping limit before spline correction. Applies only ifremove_splineisTrue. Default is3.sigma_upperThe number of standard deviations to use as the upper bound for sigma clipping limit before spline correction. Applies only ifremove_splineisTrue. Default is3.outlier_ratioMissing value threshold in % below which position correction values (POS_CORR) are used for K2SC. Missing POS_CORR values will reduce the light curve points! Otherwise, less reliable photometrically estimated centroids will be used. Default is2.THThreshold to segment each target in each TPF candence. Only used if targets cannot be separated normally. Default is8. Do not change this value unless you are aware of what you are doing!ROI_lowerThe aperture frequency grid range of interest threshold given in absolute number of selections above which pixels are considered to define the apertures. Do not change this value unless you are aware of what you are doing! Default is100.ROI_upperThe aperture frequency grid range of interest threshold given in relative number of selections w.r.t. the number of all cadences below which pixels are considered to define the apertures. Do not change this value unless you are aware of what you are doing! Default is0.85.**kwargsDictionary of arguments to be passed tok2sc.detrend.
Command-line tools
After installation, autoEAP will be available from the command line:
autoEAP <EPIC number or TPF path> [options]
Listed below are the usage instructions:
$ autoeap --help
usage: autoeap [-h] [--campaign <campaign-number>] [--applyK2SC] [--removespline] [--windowlength <window-length-in-days>] [--sigmalower <sigma-lower>] [--sigmaupper <sigma-upper>] [--saveplots]
[--TH <threshold-value>] [--ROIlower <lower-ROI-value>] [--ROIupper <upper-ROI-value>]
<path-to-targettpf-or-EPIC-number>
Perform autoEAP photometry on K2 variable stars.
positional arguments:
<path-to-targettpf-or-EPIC-number>
The location of the local TPF file or an EPIC number to be downloaded from MAST. Valid inputs include: The name of the object as a string, e.g. 'ktwo251812081'. The EPIC identifier as an integer, e.g. 251812081.
optional arguments:
-h, --help show this help message and exit
--campaign <campaign-number>
If the target has been observed in more than one campaign, download this light curve. If not given, the first campaign will be downloaded.
--applyK2SC After the raw photomery, apply K2SC to remove systematics from the extracted light curve.
--removespline After the raw or K2SC photomery, remove a low-order spline from the extracted light curve.
--windowlength <window-length-in-days>
The length of filter window for spline correction given in days. Default is 20 days.
--sigmalower <sigma-lower>
The number of standard deviations to use as the lower bound for sigma clipping limit before spline correction. Default is 3.
--sigmaupper <sigma-upper>
The number of standard deviations to use as the upper bound for sigma clipping limit before spline correction. Default is 3.
--outlierratio <outlier-ratio>
Missing value threshold in % below which position correction values (POS_CORR) are used for K2SC. Missing POS_CORR values will reduce the light curve points! Otherwise, less reliable photometrically estimated centroids will be used. Default is 2.
--saveplots Save all the plots that show each step into a subdirectory.
--TH <threshold-value>
Threshold to segment each target in each TPF candence. Only used if targets cannot be separated normally. Default is 8.
--ROIlower <lower-ROI-value>
The aperture frequency grid range of interest threshold given in absolute number of selections above which pixels are considered to define the apertures. Default is 100.
--ROIupper <upper-ROI-value>
The aperture frequency grid range of interest threshold given in relative number of selections w.r.t. the number of all cadences below which pixels are considered to define the apertures. Default is 0.85.
Data Access
We provide photometry for targets for the following Guest Observation Programs:
GO12111,GO8037,GO13111,GO14058,GO6082,GO16058,GO18033,GO10037,GO15058,GO17033.
Slightly less than 2000 RRLs. See: K2 approved targets & programs.
The data we have already created have been uploaded to our webpage.
Contributing
Feel free to open PR / Issue, or contact us here or here.
Citing
If you find this code useful, please cite Plachy et al.,2019,ApJS,244,32, until the new paper is not ready. Here is the BibTeX source:
@ARTICLE{2019ApJS..244...32P,
author = {{Plachy}, Emese and {Moln{\'a}r}, L{\'a}szl{\'o} and {B{\'o}di}, Attila and {Skarka}, Marek and {Szab{\'o}}, P{\'a}l and {Szab{\'o}}, R{\'o}bert and {Klagyivik}, P{\'e}ter and {S{\'o}dor}, {\'A}d{\'a}m and {Pope}, Benjamin J.~S.},
title = "{Extended Aperture Photometry of K2 RR Lyrae stars}",
journal = {\apjs},
keywords = {RR Lyrae variable stars: 1410, Light curves (918, Space telescopes (1547, 1410, 918, 1547, Astrophysics - Instrumentation and Methods for Astrophysics, Astrophysics - Solar and Stellar Astrophysics},
year = 2019,
month = oct,
volume = {244},
number = {2},
eid = {32},
pages = {32},
doi = {10.3847/1538-4365/ab4132},
archivePrefix = {arXiv},
eprint = {1909.00446},
primaryClass = {astro-ph.IM},
adsurl = {https://ui.adsabs.harvard.edu/abs/2019ApJS..244...32P},
adsnote = {Provided by the SAO/NASA Astrophysics Data System}
}
Acknowledgements
This project was made possible by the funding provided by the National Research, Development and Innovation Office of Hungary, funding granted under project 2018-2.1.7-UK_GYAK-2019-00009 and by the Lendület Program of the Hungarian Academy of Sciences, project No LP2018-7.
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.