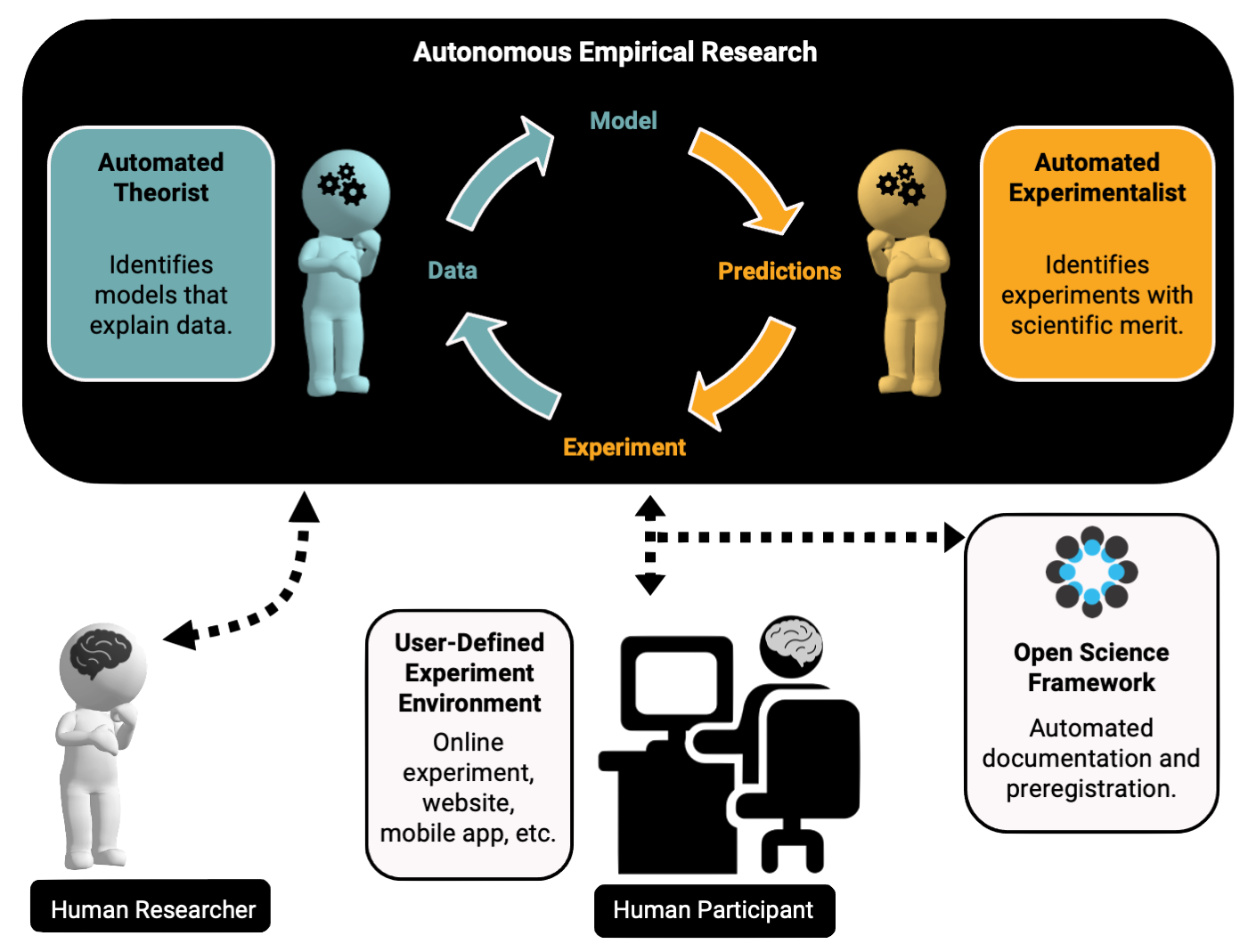
Autonomous Research Assistant (AutoRA) is a framework for automating steps of the empirical research process.
Project description
Automated Research Assistant
AutoRA (Automated Research Assistant) is an open-source framework for automating multiple stages of the empirical research process, including model discovery, experimental design, data collection, and documentation for open science.
AutoRA was initially intended for accelerating research in the behavioral and brain sciences. However, AutoRA is designed as a general framework that enables automation of the research processes in other empirical sciences, such as material science or physics.
Installation
We recommend using a Python environment manager like virtualenv. You may refer to the Development Guide on how to set up a virtual environment.
Before installing the PyPI autora package, you may activate your environment. To install the PyPI autora package, run the following command:
pip install "autora"
Documentation
Check out tutorials and documentation at https://autoresearch.github.io/autora. If you run into any issues or questions regarding the use of AutoRA, please reach out to us at the AutoRA forum.
Example
The following basic example demonstrates how to use AutoRA to automate the process of model discovery, experimental design, and data collection.
The discovery problem is defined by a single independent variable $x \in [0, 2 \pi]$ and dependent variable $y$. The experiment amounts to a simple sine wave, $y = \sin(x)$, which is the model we are trying to discover.
Th discovery cycle iterates between the experimentalist, experiment runner, and theorist. Here, we us a "random" experimentalist, which samples novel experimental conditions for $x$ every cycle. The experiment runner then collects data for the corresponding $y$ values. Finally, the theorist uses a Bayesian Machine Scientist (BMS; Guimerà et al., in Science Advances) to identify a scientific model that explains the data.
The workflow relies on the StandardState object, which stores the current state of the discovery process, such as conditions, experiment_data, or models. The state is passed between the experimentalist, experiment runner, and theorist.
####################################################################################
## Import statements
####################################################################################
import pandas as pd
import numpy as np
import sympy as sp
from autora.variable import Variable, ValueType, VariableCollection
from autora.experimentalist.random import random_pool
from autora.experiment_runner.synthetic.abstract.equation import equation_experiment
from autora.theorist.bms import BMSRegressor
from autora.state import StandardState, on_state, estimator_on_state
####################################################################################
## Define initial data
####################################################################################
#### Define variable data ####
iv = Variable(name="x", value_range=(0, 2 * np.pi), allowed_values=np.linspace(0, 2 * np.pi, 30))
dv = Variable(name="y", type=ValueType.REAL)
variables = VariableCollection(independent_variables=[iv],dependent_variables=[dv])
#### Define seed condition data ####
conditions = random_pool(variables, num_samples=10, random_state=0)
####################################################################################
## Define experimentalist
####################################################################################
experimentalist = on_state(random_pool, output=["conditions"])
####################################################################################
## Define experiment runner
####################################################################################
sin_experiment = equation_experiment(sp.simplify('sin(x)'), variables.independent_variables, variables.dependent_variables[0])
sin_runner = sin_experiment.experiment_runner
experiment_runner = on_state(sin_runner, output=["experiment_data"])
####################################################################################
## Define theorist
####################################################################################
theorist = estimator_on_state(BMSRegressor(epochs=100))
####################################################################################
## Define state
####################################################################################
s = StandardState(
variables = variables,
conditions = conditions,
experiment_data = pd.DataFrame(columns=["x","y"])
)
####################################################################################
## Cycle through the state
####################################################################################
print('Pre-Defined State:')
print(f"Number of datapoints collected: {len(s['experiment_data'])}")
print(f"Derived models: {s['models']}")
print('\n')
for i in range(5):
s = experimentalist(s, num_samples=10, random_state=42)
s = experiment_runner(s, added_noise=1.0, random_state=42)
s = theorist(s)
print(f"\nCycle {i+1} Results:")
print(f"Number of datapoints collected: {len(s['experiment_data'])}")
print(f"Derived models: {s['models']}")
print('\n')
If you are curious about how to apply AutoRA to real-world discovery problems, you can find use case examples of AutoRA in the Use Case Tutorials section of the documentation.
Contributions
We welcome contributions to the AutoRA project. Please refer to the contributor guide for more information. Also, feel free to ask any questions or provide any feedback regarding core contributions on the AutoRA forum.
About
This project is in active development by the Autonomous Empirical Research Group.
The development of this package was supported by Schmidt Science Fellows, in partnership with the Rhodes Trust, as well as the Carney BRAINSTORM program at Brown University. The development of auxiliary packages for AutoRA, such as autodoc, is supported by Schmidt Sciences, LLC. and the Virtual Institute for Scientific Software (VISS). The AutoRA package was developed using computational resources and services at the Center for Computation and Visualization at Brown University.
Read More
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
Filter files by name, interpreter, ABI, and platform.
If you're not sure about the file name format, learn more about wheel file names.
Copy a direct link to the current filters
File details
Details for the file autora-4.2.0.tar.gz.
File metadata
- Download URL: autora-4.2.0.tar.gz
- Upload date:
- Size: 7.4 MB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/6.0.1 CPython/3.9.20
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
8e7ecfac6b9f33556bf5e901ca006d35feb3106493611bdd0ea55b9a994251a8
|
|
| MD5 |
9cdafa4145534be7cceefb209feac946
|
|
| BLAKE2b-256 |
464ec64fb268183a2c25124469615b52fb9e5424f8638ebd2969c6fb1e8ad662
|
File details
Details for the file autora-4.2.0-py3-none-any.whl.
File metadata
- Download URL: autora-4.2.0-py3-none-any.whl
- Upload date:
- Size: 6.1 kB
- Tags: Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/6.0.1 CPython/3.9.20
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
c40fcf3c2f4da2c33cc67fc6b7476d3854da9286b85ae6f810e005532a93b3e4
|
|
| MD5 |
c97ece088e019f5bfe0521e9e193117a
|
|
| BLAKE2b-256 |
6d4fb2a2a9dddd2fad90204db582c06989ec709744da4036492aa5e97c1fcf9a
|