The semi-automatic segmentation and quantification of patchy areas in various biomedical images based on the assessment of their local edge densities.
Project description
BCAnalyzer: Segmentation of patchy areas in biomedical images based on local edge density estimation




Installation
Download executable from release page
We provide a builded windows executable file for every release. You can find the latest whl or exe file on the release page. Also, you can find all versions of windows execution files in the archive.
Installation from binaries
pip install -U bio-cntrs-analyzer
Build exe from source
From venv:
pip install cx_Freeze
python setup.py build
User manual
For starting appliation evualate in command line (terminal) the next command:
bcanalyzer
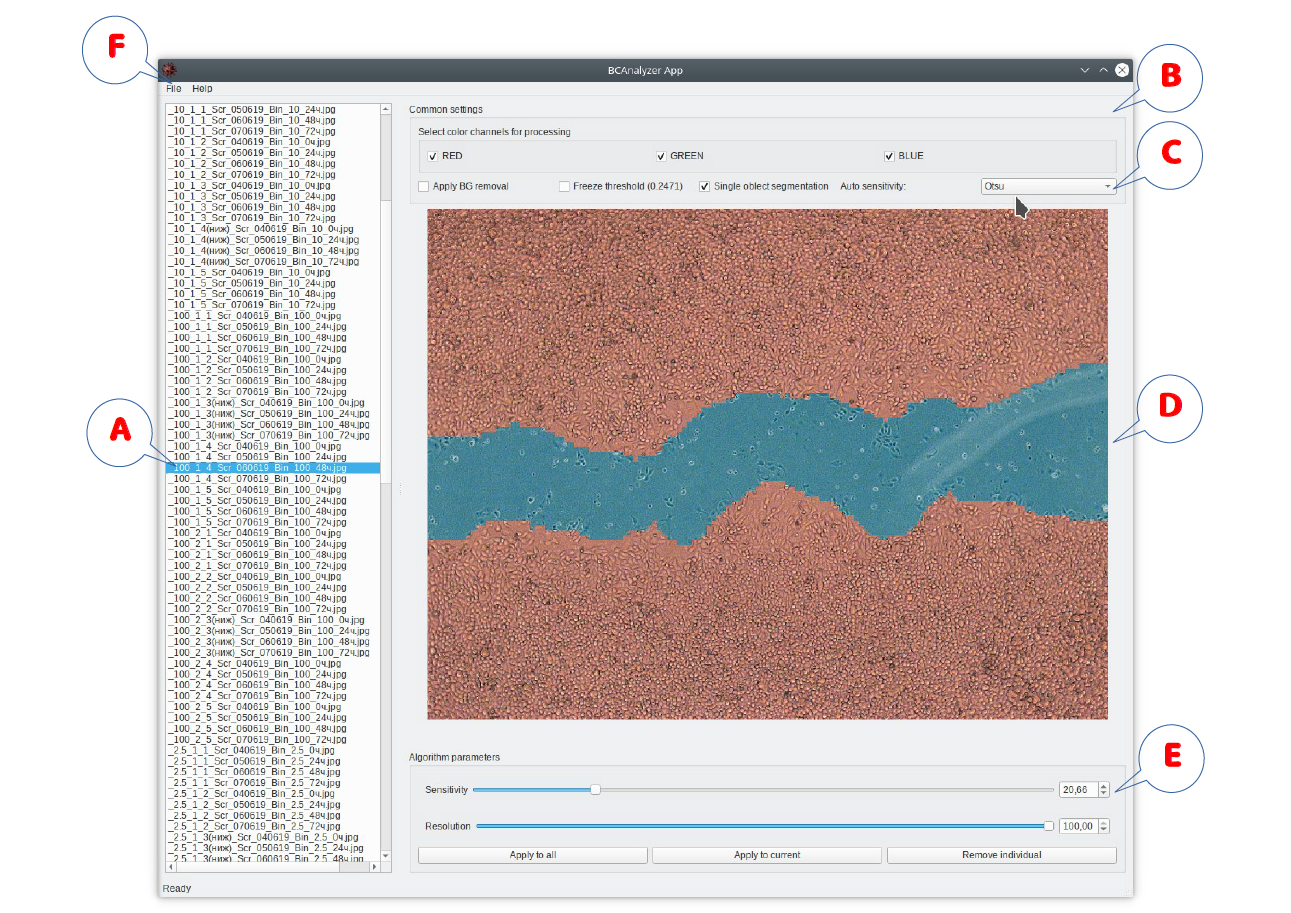
Software user interface outline: (A) list of images submitted for the analysis; (B) segmentation algorithm options, including (C) color channel import options; (D) selected image with on-the-fly visualization of the segmentation results; (E) segmentation algorithm controls for the online adjustment of its sensitivity and resolution; (F) file export menu.
The typical algorithm of user interaction with the software is as follows:
- Selected images are imported by their drag-and-drop onto the program window. The image list appears in A.
- Global algorithm options can be adjusted in B and color channels for the analysis selected in C.
- The image selected in the list A is displayed in the panel D with immediate visualization of the segmentation results (using default parameters during the first run).
- Next the algorithm parameters (sensitivity and resolution) can be adjusted manually in E, given that the automated threshold option is disabled, although one can also apply the automated threshold selection for the first approach and then disable it in order to proceed with manual fine tuning. Segmentation results are visualized on-the-fly for direct user control. Of note, global options and color channel selection can be readjusted at this stage as well. Following necessary adjustments, the chosen algorithm parameters can be applied either to the entire imported image set, or solely to the currently analyzed image, with corresponding controls available in E.
- Once the algorithm parameters are adjusted either for a single or for a few representative image(s), further processing and export can be performed as a fully automated procedure for the entire image set using file export options in F. Export options include visualizations of segmentation results, either as binary masks or as marked-up images similar to those appearing on the screen during the analysis, or both of them, as well as a *.csv table with summary statistics.
Dataset
Link to downloading dataset:
If something goes wrong, please, write to amsinitca[at]etu.ru
Troubleshooting
qt.qpa.plugin: Could not load the Qt platform plugin "xcb" in "" even though it was found.
Problem
qt.qpa.plugin: Could not load the Qt platform plugin "xcb" in "" even though it was found.
This application failed to start because no Qt platform plugin could be initialized. Reinstalling the application may fix this problem.
Available platform plugins are: eglfs, linuxfb, minimal, minimalegl, offscreen, vnc, wayland-egl, wayland, wayland-xcomposite-egl, wayland-xcomposite-glx, webgl, xcb.
Aborted (core dumped)
Solution
sudo apt-get install libxcb-xinerama0
Citation
If you find this project useful, please cite:
@article{SINITCA2023104189,
title = {Segmentation of patchy areas in biomedical images based on local edge density estimation},
journal = {Biomedical Signal Processing and Control},
volume = {79},
pages = {104189},
year = {2023},
issn = {1746-8094},
doi = {https://doi.org/10.1016/j.bspc.2022.104189},
url = {https://www.sciencedirect.com/science/article/pii/S1746809422006437},
author = {Aleksandr M. Sinitca and Airat R. Kayumov and Pavel V. Zelenikhin and Andrey G. Porfiriev and Dmitrii I. Kaplun and Mikhail I. Bogachev},
}
Project details
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
File details
Details for the file Bio-Cntrs-Analyzer-0.2.0.tar.gz.
File metadata
- Download URL: Bio-Cntrs-Analyzer-0.2.0.tar.gz
- Upload date:
- Size: 149.6 kB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: poetry/1.2.1 CPython/3.10.4 Windows/10
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | e14c512e36783c82a0097169d3f5dfe048827877a49b2dd5825ce16e22582420 |
|
| MD5 | cd14170e8cd9a2606cf5ad7f93b621ed |
|
| BLAKE2b-256 | e44e33e927dc01f0f60054fe7a379e0a91f41437dd0ba2ba75b167d531e35abe |
File details
Details for the file bio_cntrs_analyzer-0.2.0-py3-none-any.whl.
File metadata
- Download URL: bio_cntrs_analyzer-0.2.0-py3-none-any.whl
- Upload date:
- Size: 152.2 kB
- Tags: Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: poetry/1.2.1 CPython/3.10.4 Windows/10
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | ffafe88f1da633f128e9a5ac8d8c9dfb49a91d9dfcdd77135725d5d14ca30947 |
|
| MD5 | e7a60262cf4855351d44d8b61c6ccc3d |
|
| BLAKE2b-256 | 9fcb1bc5aca317b6b4951f43eba5fdf03b3513eb7204c8ea882843fd1cbb47ec |