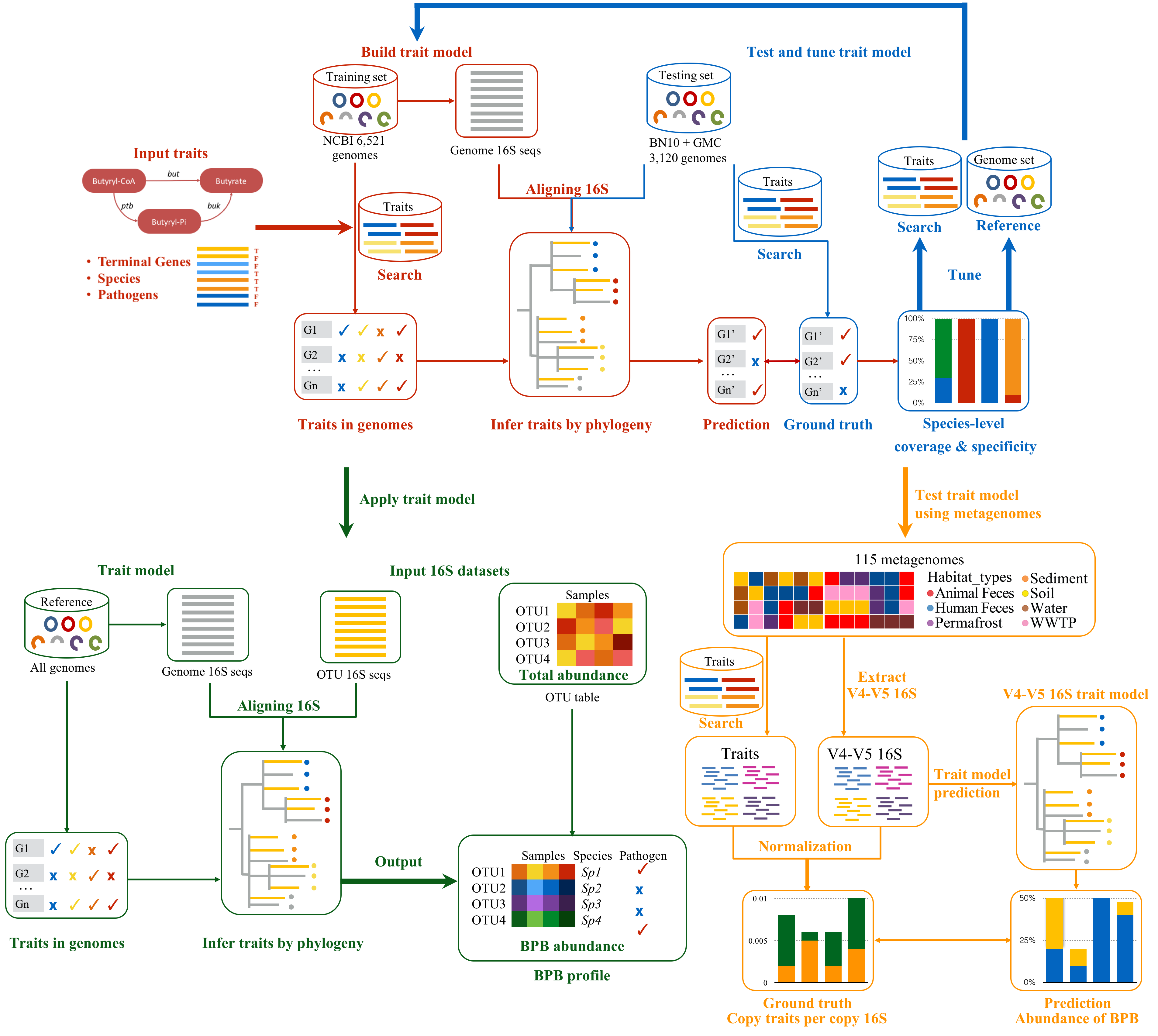
Inferring traits by 16S
Project description
buty_phyl
Introduction
- buty_phyl infers traits by 16S
- input: otu table (-t) and otu sequences (-s)
- requirement: mafft
- Optional: fasttree
Install
pip install buty_phyl
in preparation: anaconda download caozhichongchong/buty_phyl
Availability
in preparation: https://anaconda.org/caozhichongchong/buty_phyl
https://pypi.org/project/buty_phyl
How to use it
-
test the buty_phyl
buty_phyl --test -
try your data
buty_phyl -t your.otu.table -s your.otu.seqs
buty_phyl -t your.otu.table -s your.otu.seqs -top 2000 -
try different traits (default is butyrate production)
predicting butyrate production
buty_phyl -t your.otu.table -s your.otu.seqs
or
buty_phyl -t your.otu.table -s your.otu.seqs --rt b
predicting sulfate reduction
buty_phyl -t your.otu.table -s your.otu.seqs --rt s
predicting nitrate reduction
buty_phyl -t your.otu.table -s your.otu.seqs --rt n -
use your own traits
buty_phyl -t your.otu.table -s your.otu.seqs --rs your.own.reference.16s --rt your.own.reference.traits
-
your.own.reference.16s is a fasta file containing the 16S sequences of your genomes
>Genome_ID1
ATGC...
>Genome_ID2
ATGC... -
your.own.reference.traits is a metadata of whether there's trait in your genomes (0 for no and 1 for yes)
Genome_ID1 0
Genome_ID1 1
Results
The result dir of "Bayers_model":
filename.infertraits.txt: the OTUs inferring as butyrate-producing bacteria (1.0), unknown bacteria (0.5), and non-butyrate-producing bacteria (0.0).filename.infertraits.abu: the total abundance of butyrate-producing bacteria.filename.infertraits.commensal.abu: the total abundance of commensal butyrate-producing bacteria.filename.infertraits.pathogen.abu: the total abundance of pathogenic butyrate-producing bacteria.filename.infertraits.otu_table: the otu_table of butyrate-producing bacteria.filename.infertraits.commensal.otu_table: the otu_table of commensal butyrate-producing bacteria.filename.infertraits.pathogen.otu_table: the otu_table of pathogenic butyrate-producing bacteria.filename.bpbspecies.commensal.abu: the abundance of commensal butyrate-producing species.filename.bpbspecies.pathogen.abu: the abundance of pathogenic butyrate-producing species.
The result dir of "Filtered_OTU":
- Some temp files of filtered OTUs, alignment, and tree.
Copyright
Copyright: An Ni Zhang, Prof. Eric Alm, Alm Lab in MIT
Citation: Not yet, coming soon!
Contact: anniz44@mit.edu
Project details
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
Filter files by name, interpreter, ABI, and platform.
If you're not sure about the file name format, learn more about wheel file names.
Copy a direct link to the current filters
File details
Details for the file buty_phyl-1.0.7.tar.gz.
File metadata
- Download URL: buty_phyl-1.0.7.tar.gz
- Upload date:
- Size: 4.7 MB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/1.12.1 pkginfo/1.5.0.1 requests/2.22.0 setuptools/41.0.1 requests-toolbelt/0.8.0 tqdm/4.32.2 CPython/3.6.7
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
2189c3c5830ab13f64da329ace83052882bbe870228809a30487e21e3b55dfeb
|
|
| MD5 |
3ceb87f32b943afc1e74edda88478d88
|
|
| BLAKE2b-256 |
4e7fccd73b54710b8d0df6214ad922d2f11f7b27de23df5980a4fc820f3b067b
|
File details
Details for the file buty_phyl-1.0.7-py3.6.egg.
File metadata
- Download URL: buty_phyl-1.0.7-py3.6.egg
- Upload date:
- Size: 5.7 MB
- Tags: Egg
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/1.12.1 pkginfo/1.5.0.1 requests/2.22.0 setuptools/41.0.1 requests-toolbelt/0.8.0 tqdm/4.32.2 CPython/3.6.7
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
4103ea625fd80cba8d399e63810bcd374c5cbed0e4dba0bcc72465b107b7d2cc
|
|
| MD5 |
006b243b1fe04304312c20b53477d9f6
|
|
| BLAKE2b-256 |
2c0a479f1ea234e8e488a0b5df0cfff223411356a7f7355de016a83ca441830c
|