This project provides a structure to integrate calcium traces and events, perform statistical analysis, make graphs and do classification problems to further understand the dataset.
Project description
Calcium Imaging Data Analysis
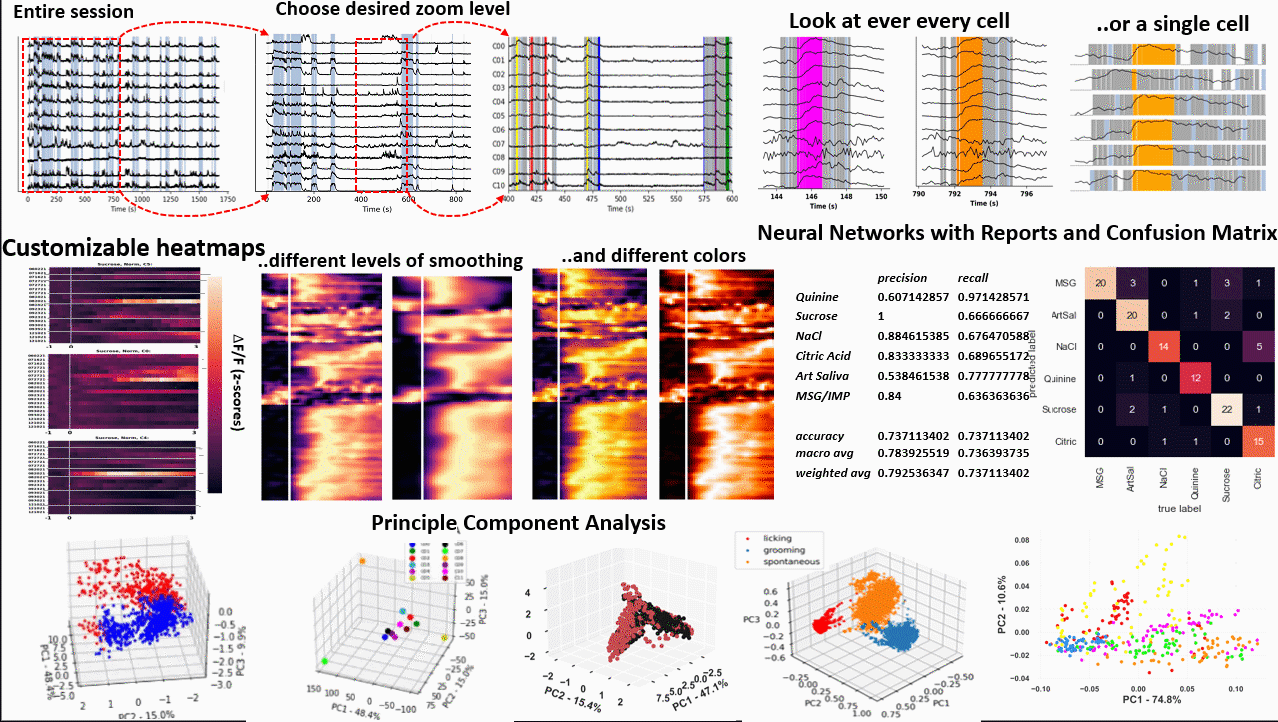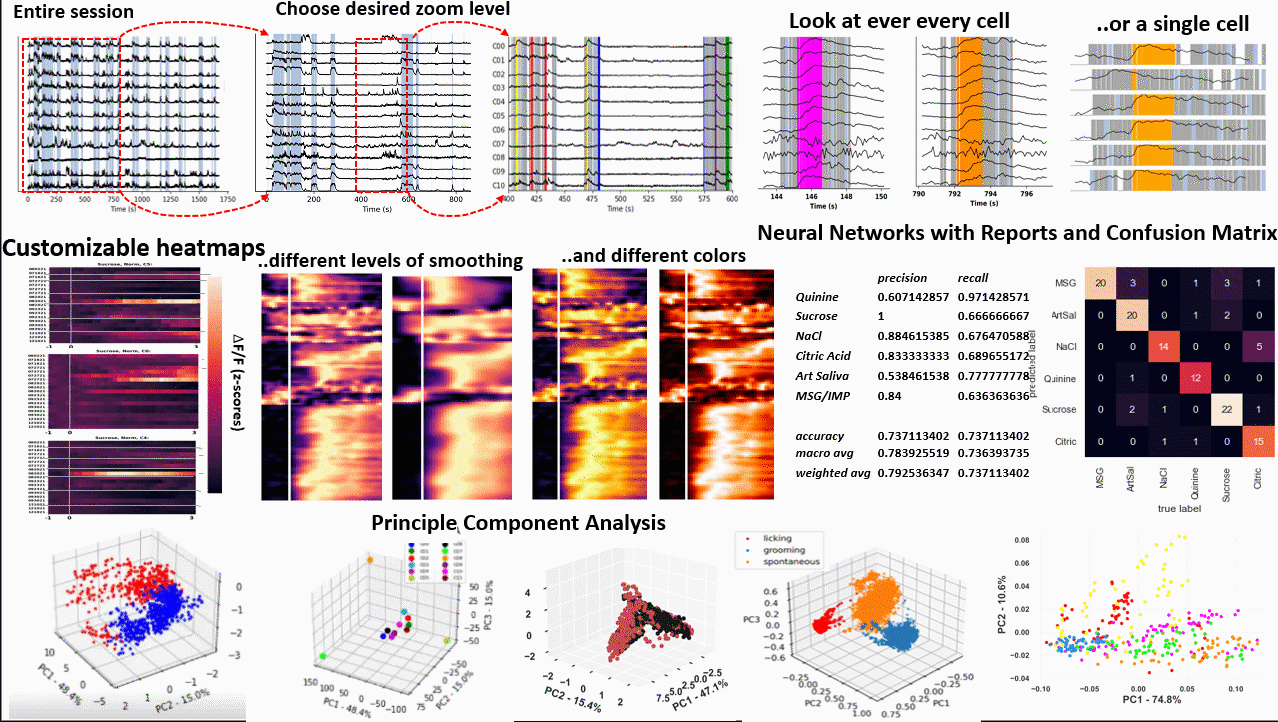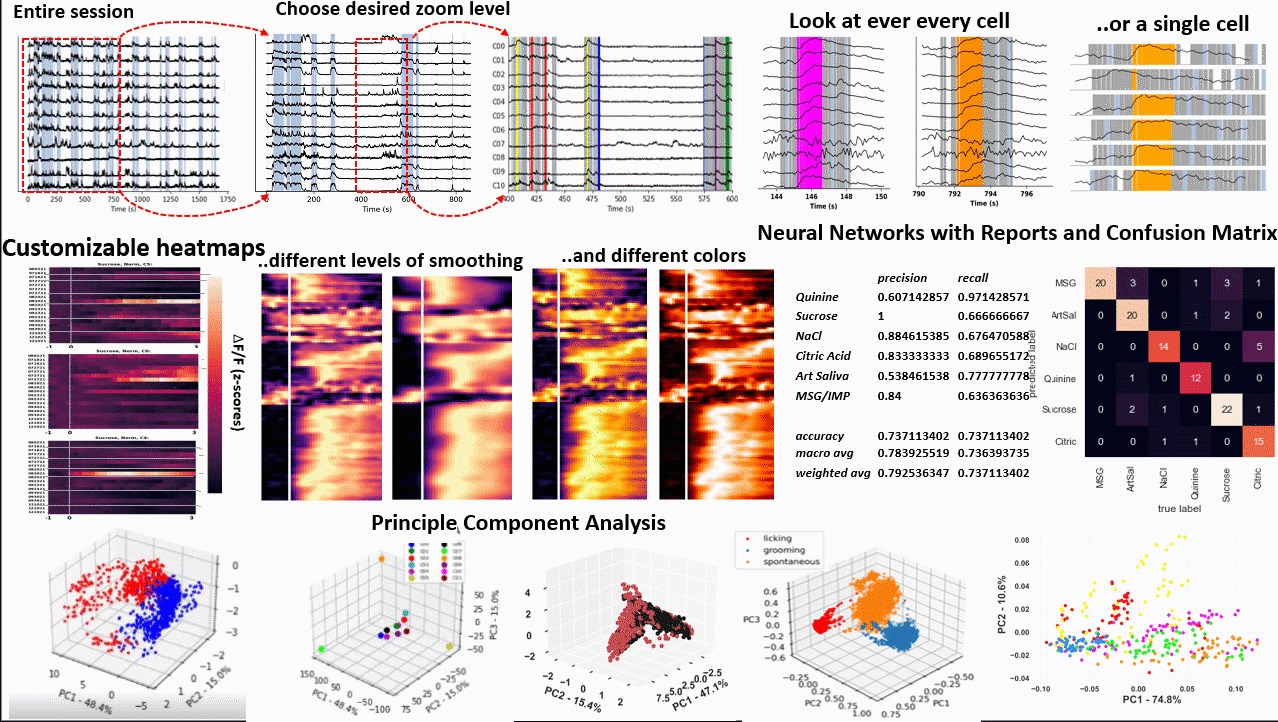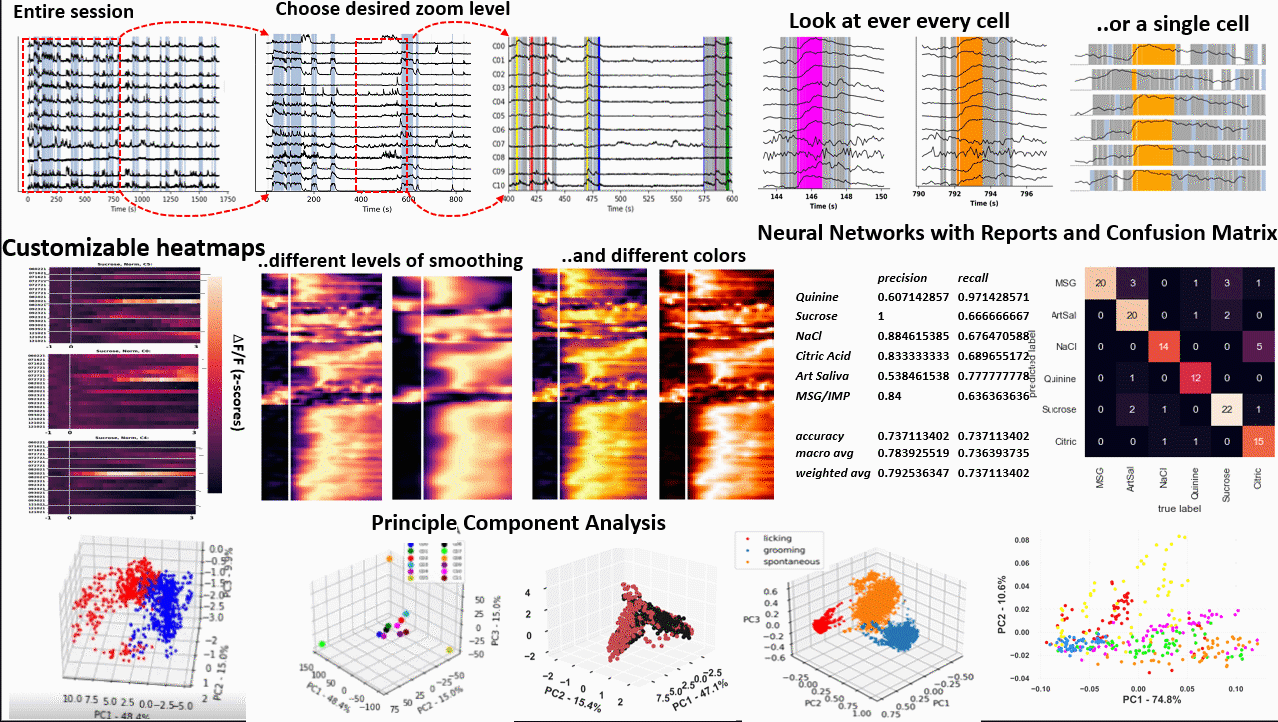
*Images from poster presented at AChems 2022 [1].
This project takes .csv files gathered from Inscopix Data Processing software for processing:
- Syncing traces with externally captured GPIO events.
- General
based statistics.
- Plotting: animated, 2D and 3D scatter, regression, skree, heatmap and correlation matrix.
- Dimensionality reduction with variance filters and principal component analysis.
- Support Vector Machine Learning for classification tasks.
| Supported OS | Linux (list of POSIX API), Windows 7 and higher. |
Directory Structure
For data loading, directories should be formatted a particular way that can be easily
modified by configuring data.data_utils.file_handler.
./data/
├── Animal 1
│ ├── Day 1
│ │ ├── *Events.csv
│ │ └── *Traces.csv
│ └── Day 2
│ ├── *Events.csv
│ └── *Traces.csv
├── Animal 2
│ └── Day 1
│ ├── *Events.csv
│ └── *Traces.csv
└── Animal 3
└── Day 1
├── *Events.csv
└── *Traces.csv
Parameters and Directories
A configuration file config.yaml contains defaults that can be set in a single top-level file.
-
DIRS:- directories for loading and saving dataDIRS: data_dir: '/Users/me/mydata' # os is internally handled save_stats: '~./home' # save statistical output save_figs: '~/graphs' # can have multiple sub-dirs save_nn: '~/svm' # train/test/eval scores from neural network
-
SESSION:- session instance informationSESSION: animal: 'animal_ID' # must include char + num date: '123456' # must include all nun dir: '/my/data' # must include / or \
-
COLORS:- event : color pairingCOLORS: Event1: 'color' Event2: 'color' Event3: 'color' # from matplotlib.colors API
-
DECODER:- Decode GPIO signals into eventsDECODER: Event1: 1, 2, 3, 4 Event2: 1, 3, 4 Event3: 1, 2, 3 Event4: 1, 4 # see data/data_utils/gpio_data.py for implementation
TO DO:
- Additional layers to neural network for additional feature extraction
- Layer for additional outlier/anomaly detection
- Layer for linear trend normalization
- Any additional feature extraction layers i.e. temporal coding [2], latency, duration, etc.
- Incorperate dataflow-based modeling design i.e. mapping actors to threads and the vectorization of actors (see [3])
- Format all docstrings to NumPy style
References
[1] Long Term In-Vivo One-Photon Ca2+ Imaging of Taste-Responsive Cells In The Parabrachial Pons In The Freely Licking Rat. Flynn P. O'Connell1, Joshua D. Sammons2, Patricia M. Di Lorenzo1. 1Binghamton University, Binghamton, NY, USA. 2University of Alabama at Birmingham, Birmingham, AL, USA. Achems.org
[2] Hallock R. M., Di Lorenzo P. M. (2006). Temporal coding in the gustatory system. Neurosci. Biobehav. Rev. 30, 1145–1160. 10.1016/j.neubiorev.2006.07.005
[3] Kyunghun Lee, Xiaomin Wu, Yaesop Lee, Da-Ting Lin, Shuvra S. Bhattacharyya & Rong Chen (2021) Neural decoding on imbalanced calcium imaging data with a network of support vector machines, Advanced Robotics, 35:7, 459-470, DOI: 10.1080/01691864.2020.1863259
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
Filter files by name, interpreter, ABI, and platform.
If you're not sure about the file name format, learn more about wheel file names.
Copy a direct link to the current filters
File details
Details for the file canalysis-1.0.0.tar.gz.
File metadata
- Download URL: canalysis-1.0.0.tar.gz
- Upload date:
- Size: 43.4 kB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/4.0.1 CPython/3.8.13
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
b857d532af927df7ebea369dabf41535885d4d08d7c6d0f1a7f70523e0e81aa2
|
|
| MD5 |
e674eef05d3a58d50c8ee4475dec660e
|
|
| BLAKE2b-256 |
7fc96fdabef71984527d7082e889d873a2387db85945f88af86b87ef2c45afb4
|
File details
Details for the file canalysis-1.0.0-py3-none-any.whl.
File metadata
- Download URL: canalysis-1.0.0-py3-none-any.whl
- Upload date:
- Size: 52.4 kB
- Tags: Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/4.0.1 CPython/3.8.13
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
c71fc90fa65c167f2a37649175cf53503b4538ff2a257ab6ad4937faeffc0090
|
|
| MD5 |
0703a879f492d7b8d513e6ca78479dfd
|
|
| BLAKE2b-256 |
b31f2be8fb4f48f530eda4e2c75b87a691fd93f359256eeb1e96015accbf72e6
|


















