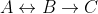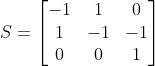A Python package to simulate biochemical systems using the Constraint-Based Simulation Algorithm
Project description
Constraint-Based Simulation Algorithm (CBSA)
CBSA is a Python package for simulation of biochemical systems using the Constraint-Based Simulation Algorithm.
Table of contents
Installation
CBSA can be installed on your computer using pip.
Just run the following command:
python3 -m pip install cbsa --user --upgrade
Usage
CBSA
Simple reaction example
Consider the following reaction system:
Using the Constrain-Based Modeling, the Stoichiometric matrix becomes:
A sample code to simulate this system is:
import cbsa
import matplotlib.pyplot as plt
import numpy as np
S = [[-1, 1, 0],
[ 1,-1,-1],
[ 0, 0, 1]]
init_mols = [50,0,0]
k = [0.5,0.1,0.8]
max_dt = 0.1
total_sim_time = 10
sim = cbsa.ReactionSystem(S)
sim.setup()
sim.set_x(init_mols)
sim.set_k(k)
sim.set_max_dt(max_dt)
sim.setup_simulation()
sim.compute_simulation(total_sim_time)
sim_data = np.array(sim.simulation_data)
plt.plot(sim_data[:,0],sim_data[:,1],label="A")
plt.plot(sim_data[:,0],sim_data[:,2],label="B")
plt.plot(sim_data[:,0],sim_data[:,3],label="C")
plt.legend()
plt.show()
License
CBSA is licensed under the MIT License. Please see the file LICENCE for more information.
Project details
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
Filter files by name, interpreter, ABI, and platform.
If you're not sure about the file name format, learn more about wheel file names.
Copy a direct link to the current filters
File details
Details for the file cbsa-0.1.0.tar.gz.
File metadata
- Download URL: cbsa-0.1.0.tar.gz
- Upload date:
- Size: 7.8 kB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/3.1.1 pkginfo/1.5.0.1 requests/2.22.0 setuptools/42.0.2 requests-toolbelt/0.9.1 tqdm/4.40.2 CPython/3.7.4
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
37bc4214d8694950efb32da6e62c9be7f6ee8ab26cff881da90f726325cbb099
|
|
| MD5 |
5cde0287fb679af6bcecf68633054ab8
|
|
| BLAKE2b-256 |
2a22fd5b3fa34f9464658f8dfb2f649200722f1adffee46d0d29951ad5ee2f99
|
File details
Details for the file cbsa-0.1.0-py3-none-any.whl.
File metadata
- Download URL: cbsa-0.1.0-py3-none-any.whl
- Upload date:
- Size: 13.6 kB
- Tags: Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/3.1.1 pkginfo/1.5.0.1 requests/2.22.0 setuptools/42.0.2 requests-toolbelt/0.9.1 tqdm/4.40.2 CPython/3.7.4
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
a33369512226727edc8469f87a492b77767191517174c4a0559d1eb0d7e98115
|
|
| MD5 |
8c18a3ea0799ad3d8aff15ae2755e475
|
|
| BLAKE2b-256 |
10865b81defa4c648d2c7938856ccf78eba528c1b4f5d4c4bdc72ee765d23097
|