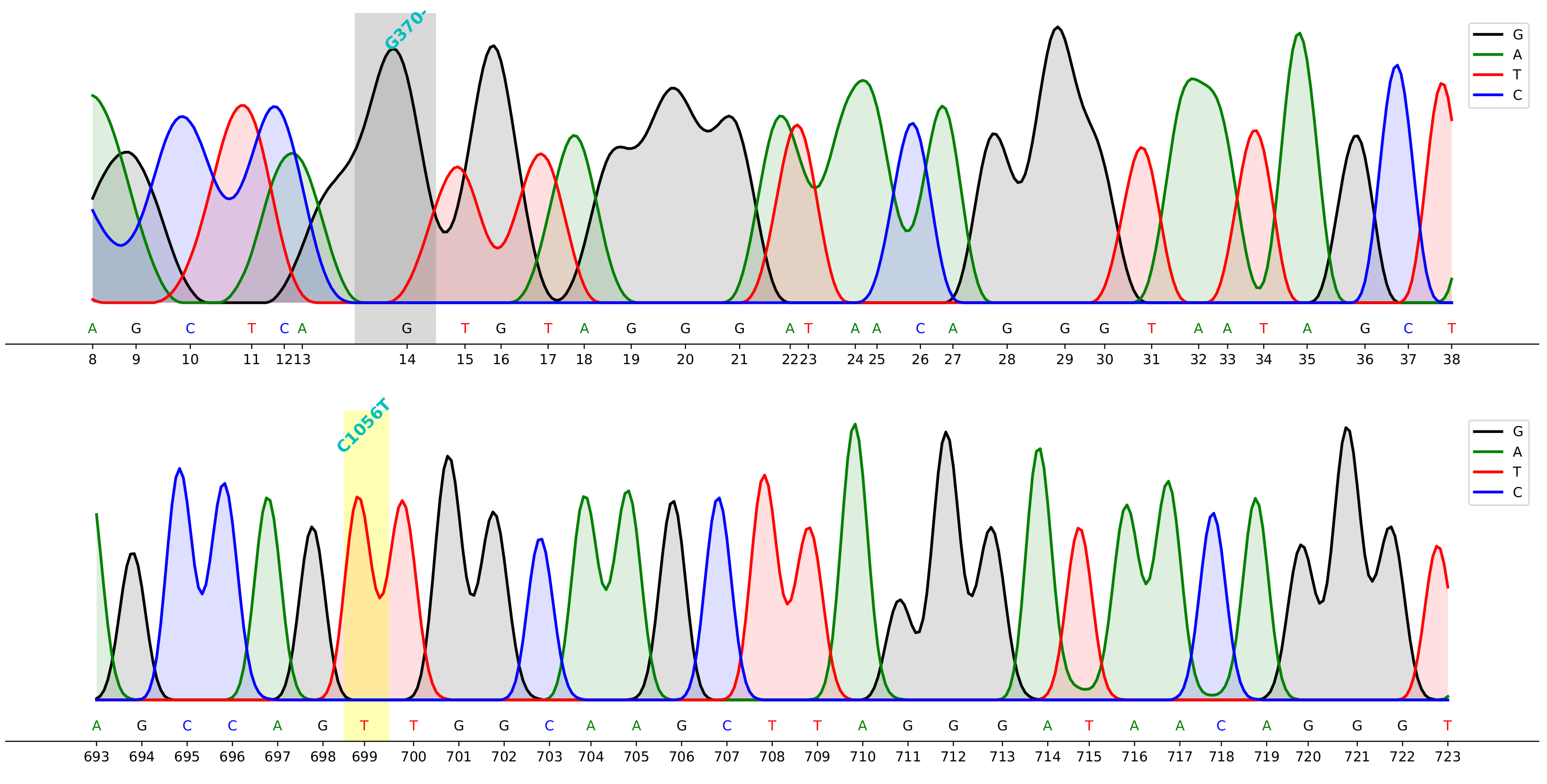
Chromatogram File Utils
Project description
Chromatogram File Utils
For Sanger sequencing data visualizing, alignment, mutation calling, and trimming etc.
Demo
command to generate the demo above
cfutils mut --query ./data/B5-M13R_B07.ab1 --subject ./data/ref.fa --outdir ./data/ --plot
How to use?
- You can have mutation detection and visualization in one step using the command line.
cfutils mut --help
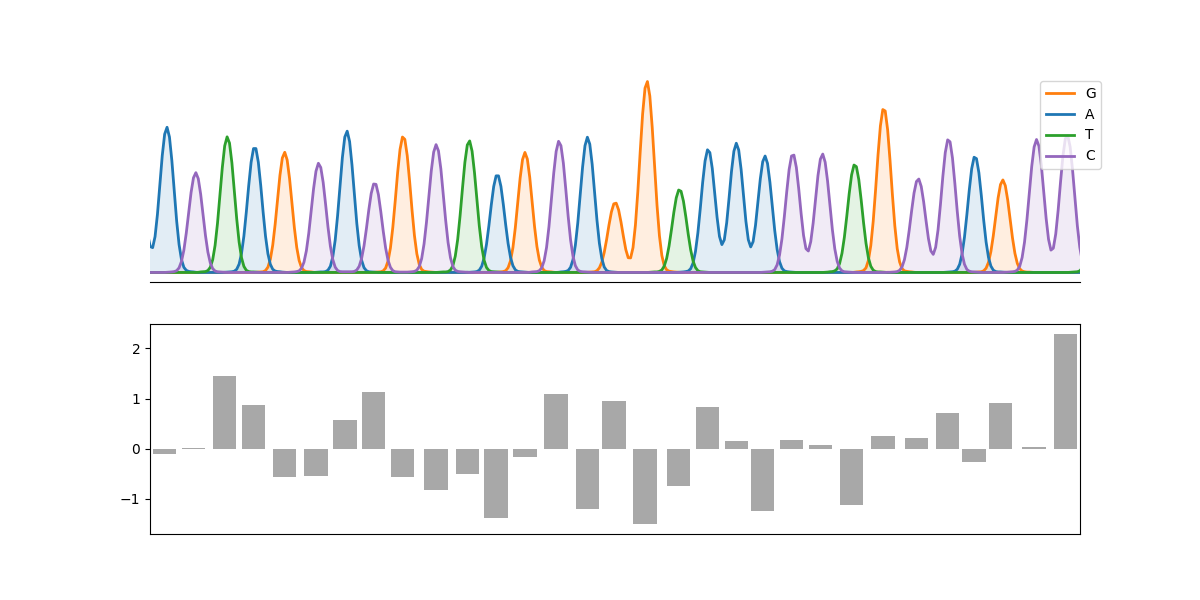
- You can also integrate the result matplotlib figures and use it as a python module.
An example:
import matplotlib.pyplot as plt
import numpy as np
from cfutils.parser import parse_abi
from cfutils.show import plot_chromatograph
seq = parse_abi("./data/B5-M13R_B07.ab1")
peaks = seq.annotations["peak positions"][100:131]
fig, axes = plt.subplots(2, 1, figsize=(12, 6), sharex=True)
plot_chromatograph(
seq,
region=(100, 130),
ax=axes[0],
show_bases=True,
show_positions=True,
color_map=dict(zip("ATGC", ["C0", "C2", "C1", "C4"])),
)
axes[1].bar(peaks, np.random.randn(len(peaks)), color="0.66")
plt.show()
How to install?
form pypi
(use this way ONLY, if you don't know what's going on)
pip install --user cfutils
manipulate the source code
- clone from github
git clone git@github.com:y9c/cfutils.git
- install the dependence
make init
- do unittest
make test
ChangeLog
- Reverse completement the chromatogram file. (Inspired by Snapgene)
- build as python package for pypi
- fix bug that highlighting wrong base
- replace blastn with buildin python aligner
TODO
-
call mutation by alignment and plot Chromatogram graphic
-
add a doc
-
change xaxis by peak location
-
fix bug that chromatogram switch pos after trim
-
wrap as a cli app
-
return quality score in output
-
fix issue that selected base is not in the middle
-
fix plot_chromatograph rendering bug
-
add projection feature to make align and assemble possible
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
Filter files by name, interpreter, ABI, and platform.
If you're not sure about the file name format, learn more about wheel file names.
Copy a direct link to the current filters
File details
Details for the file cfutils-0.0.0.dev61.tar.gz.
File metadata
- Download URL: cfutils-0.0.0.dev61.tar.gz
- Upload date:
- Size: 13.7 kB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: poetry/1.8.4 CPython/3.13.0 Darwin/23.6.0
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
196793530a94c2379297f4ee089ace09246ec9f04fafbe82e7944963da28b99d
|
|
| MD5 |
79c06f6618a88c5a25ec408da6daaafc
|
|
| BLAKE2b-256 |
9e58d477cc995a92b699a2c517472aa59f3645ae00ae1cbaf86fc50bbd3382a3
|
File details
Details for the file cfutils-0.0.0.dev61-py3-none-any.whl.
File metadata
- Download URL: cfutils-0.0.0.dev61-py3-none-any.whl
- Upload date:
- Size: 15.7 kB
- Tags: Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: poetry/1.8.4 CPython/3.13.0 Darwin/23.6.0
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
fe5a65e05fe7434a9edc15407f4a0b1f2510ced5637a6c3c02edfbbc1e46a1c7
|
|
| MD5 |
d6cd7b99bc4f34ea8e778cca17f4d54f
|
|
| BLAKE2b-256 |
56db4864c5665632a168c15fa255ff59c3a54de733ef1d7bfd080bbe5d30a3f2
|