Accelerate data driven research in developmental biology with deep learning models
Project description

Contents
- Example notebooks
- Segmenting the C. elegans embryo
- Generating synthetic images of embryos with a GAN
- Predicting populations of cells within the C. elegans embryo
- Contributing to DevoLearn
- Contact us
Installation
pip install devolearn
Example notebooks
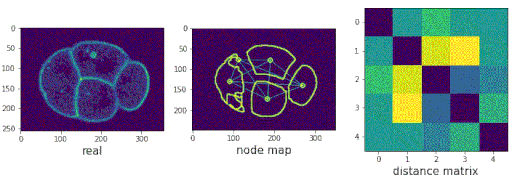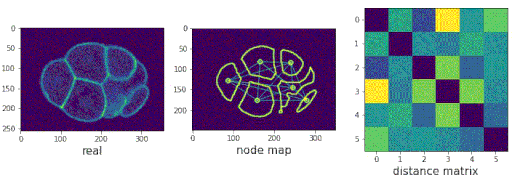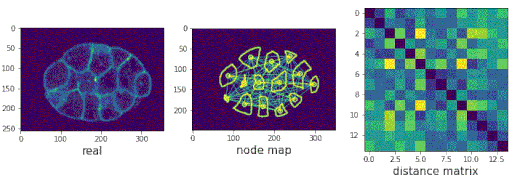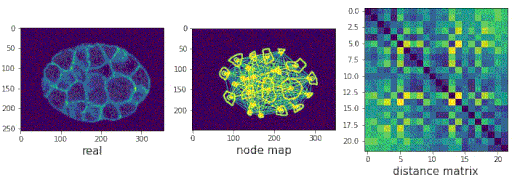
Segmenting the C. elegans embryo
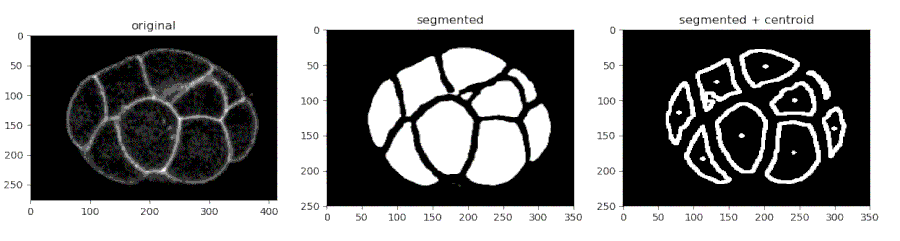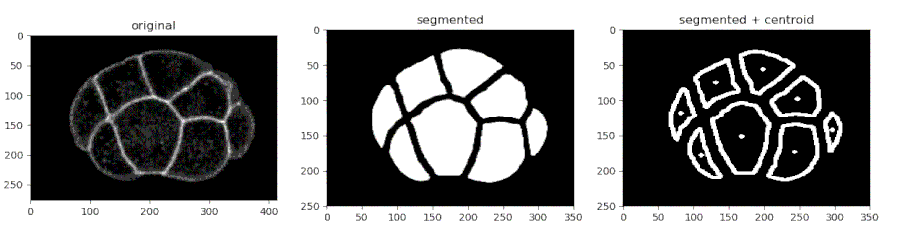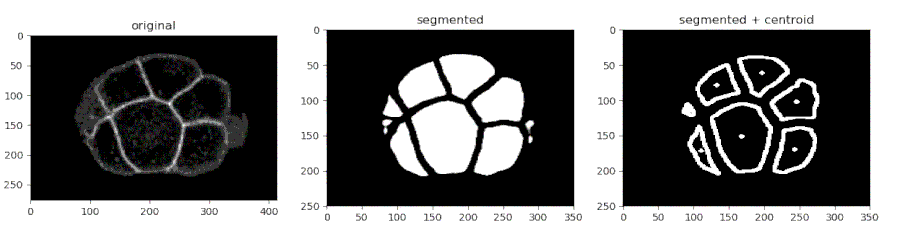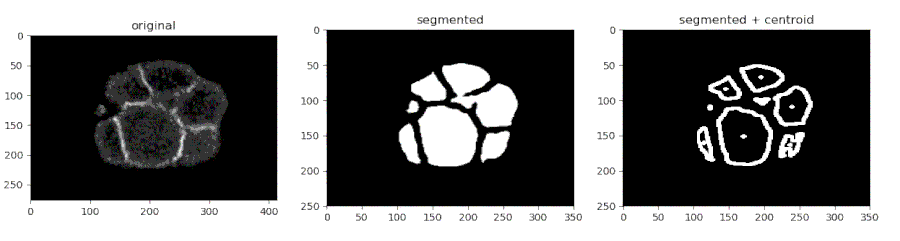
- Importing the model
from devolearn import embryo_segmentor
segmentor = embryo_segmentor()
- Running the model on an image and viewing the prediction
seg_pred = segmentor.predict(image_path = "sample_data/images/seg_sample.jpg")
plt.imshow(seg_pred)
plt.show()
- Running the model on a video and saving the predictions into a folder
filenames = segmentor.predict_from_video(video_path = "sample_data/videos/seg_sample.mov", centroid_mode = False, save_folder = "preds")
- Finding the centroids of the segmented features
seg_pred, centroids = segmentor.predict(image_path = "sample_data/images/seg_sample.jpg", centroid_mode = True)
plt.imshow(seg_pred)
plt.show()
- Saving the centroids from each frame into a CSV
df = segmentor.predict_from_video(video_path = "sample_data/videos/seg_sample.mov", centroid_mode = True, save_folder = "preds")
df.to_csv("centroids.csv")
Generating synthetic images of embryos with a Pre-trained GAN
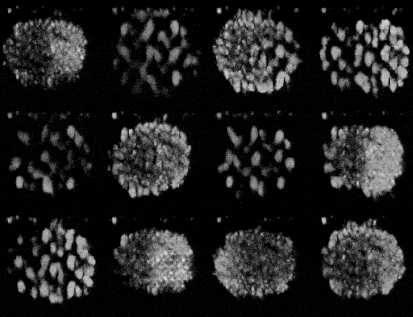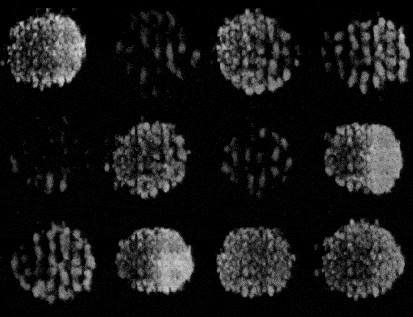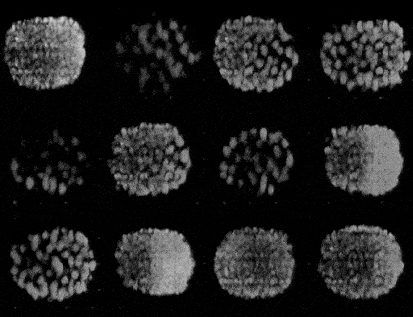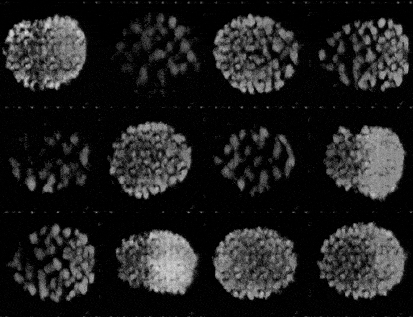
- Importing the model
from devolearn import Generator, embryo_generator_model
generator = embryo_generator_model()
- Generating a picture and viewing it with matplotlib
gen_image = generator.generate()
plt.imshow(gen_image)
plt.show()
- Generating n images and saving them into
foldernamewith a custom size
generator.generate_n_images(n = 5, foldername= "generated_images", image_size= (700,500))
Predicting populations of cells within the C. elegans embryo
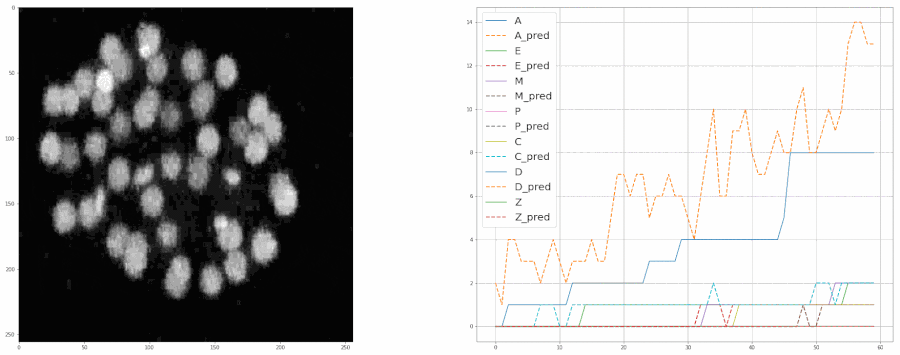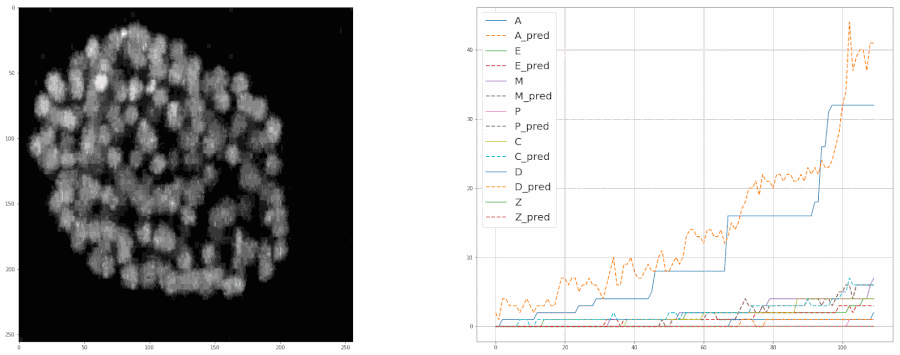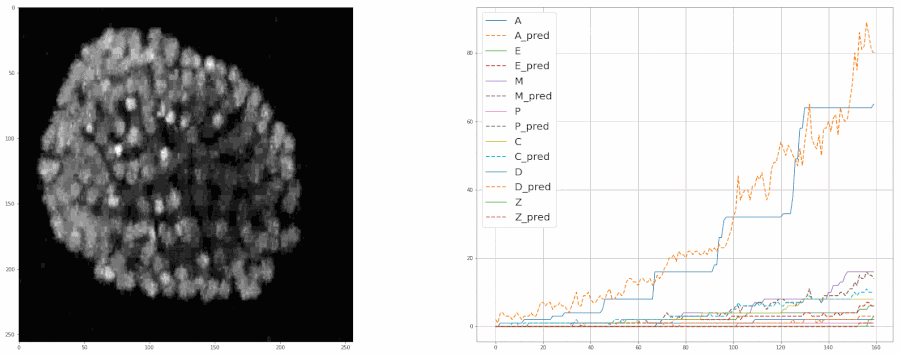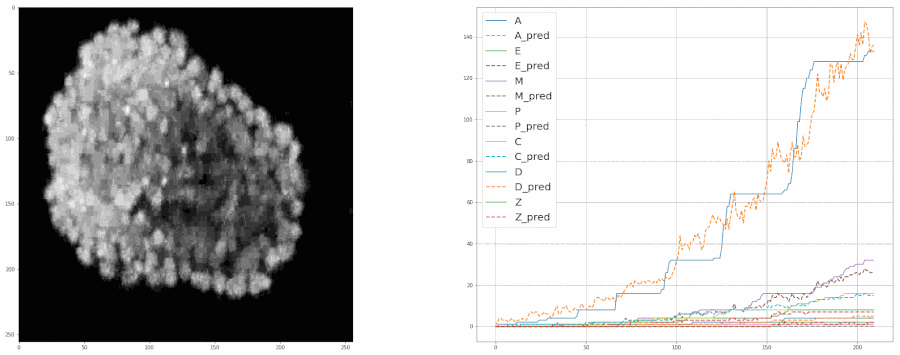
- Importing the population model for inferences
from devolearn import lineage_population_model
- Loading a model instance to be used to estimate lineage populations of embryos from videos/photos.
model = lineage_population_model(mode = "cpu")
- Making a prediction from an image
print(model.predict(image_path = "sample_data/images/embryo_sample.png"))
- Making predictions from a video and saving the predictions into a CSV file
results = model.predict_from_video(video_path = "sample_data/videos/embryo_timelapse.mov", save_csv = True, csv_name = "video_preds.csv", ignore_first_n_frames= 10, ignore_last_n_frames= 10 )
- Plotting the model's predictions from a video
plot = model.create_population_plot_from_video(video_path = "sample_data/videos/embryo_timelapse.mov", save_plot= True, plot_name= "plot.png", ignore_last_n_frames= 0 )
plot.show()
Links to Datasets
| Model | Data source |
|---|---|
| Segmenting the C. elegans embryo | 3DMMS: robust 3D Membrane Morphological Segmentation of C. elegans embryo |
| Cell lineage population prediction + embryo GAN | EPIC dataset |
Authors/maintainers:
Feel free to join our Slack workspace!
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
devolearn-0.3.0.tar.gz
(1.9 MB
view details)
Built Distribution
Filter files by name, interpreter, ABI, and platform.
If you're not sure about the file name format, learn more about wheel file names.
Copy a direct link to the current filters
File details
Details for the file devolearn-0.3.0.tar.gz.
File metadata
- Download URL: devolearn-0.3.0.tar.gz
- Upload date:
- Size: 1.9 MB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/3.3.0 pkginfo/1.7.0 requests/2.25.1 setuptools/53.0.0 requests-toolbelt/0.9.1 tqdm/4.56.0 CPython/3.8.5
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
c41bd25f208cf1e0b3f6ea896dcc03cfc251b866c1fc6003ae884e434c308d28
|
|
| MD5 |
0a869eeb5dbc32fb3a4e76524c6f562d
|
|
| BLAKE2b-256 |
47ced07bb787ad37ea1ac301c4ec88c649a0cf3ddf0d8acfd6266c8ba76841aa
|
File details
Details for the file devolearn-0.3.0-py3-none-any.whl.
File metadata
- Download URL: devolearn-0.3.0-py3-none-any.whl
- Upload date:
- Size: 1.9 MB
- Tags: Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/3.3.0 pkginfo/1.7.0 requests/2.25.1 setuptools/53.0.0 requests-toolbelt/0.9.1 tqdm/4.56.0 CPython/3.8.5
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
c62c1382071cb61c84cae1806ecb0843704e8348cf7c6b2eefa1e149defc4efd
|
|
| MD5 |
c529783fe98e3bc2ba0e62f481a12a2e
|
|
| BLAKE2b-256 |
cb35ef245a00a00632ea0a2e6c4c58e863755d4681497ce5dff0209241297f52
|

















