PyMOL plugin to interactively label nucleic acids with fluorophores in silico
Project description

FRETlabel is a PyMOL plugin to label nucleic acids with explicit fluorescent dyes. It aims to facilitate the workflow of setting up, running and evaluating molecular dynamics simulations with explicit organic fluorophores for in silico FRET calculations.
Specifically, FRETlabel includes the following features:
- PyMOL plugin for fluorescent labeling: Label your nucleic acid of interest with the click of a button. The PyMOL plugin extends the AMBERDYES package (Graen et al. JCTC 2014) geometries and force field parameters of common nucleic acid linker chemistries.
- Build new fragments interactively: Tutorials guide you step-by-step through the process of creating new base, linker and dye fragments by integrating with established pipelines for topology generation such as Antechamber and Acpype.
- FRET prediction: Calculate FRET distributions from MD simulation with all-atom organic dyes. FRETlabel integrates with FRETraj to compute photon bursts based on distance RDA(t) and orientation trajectories κ2(t) of the fluorophores.
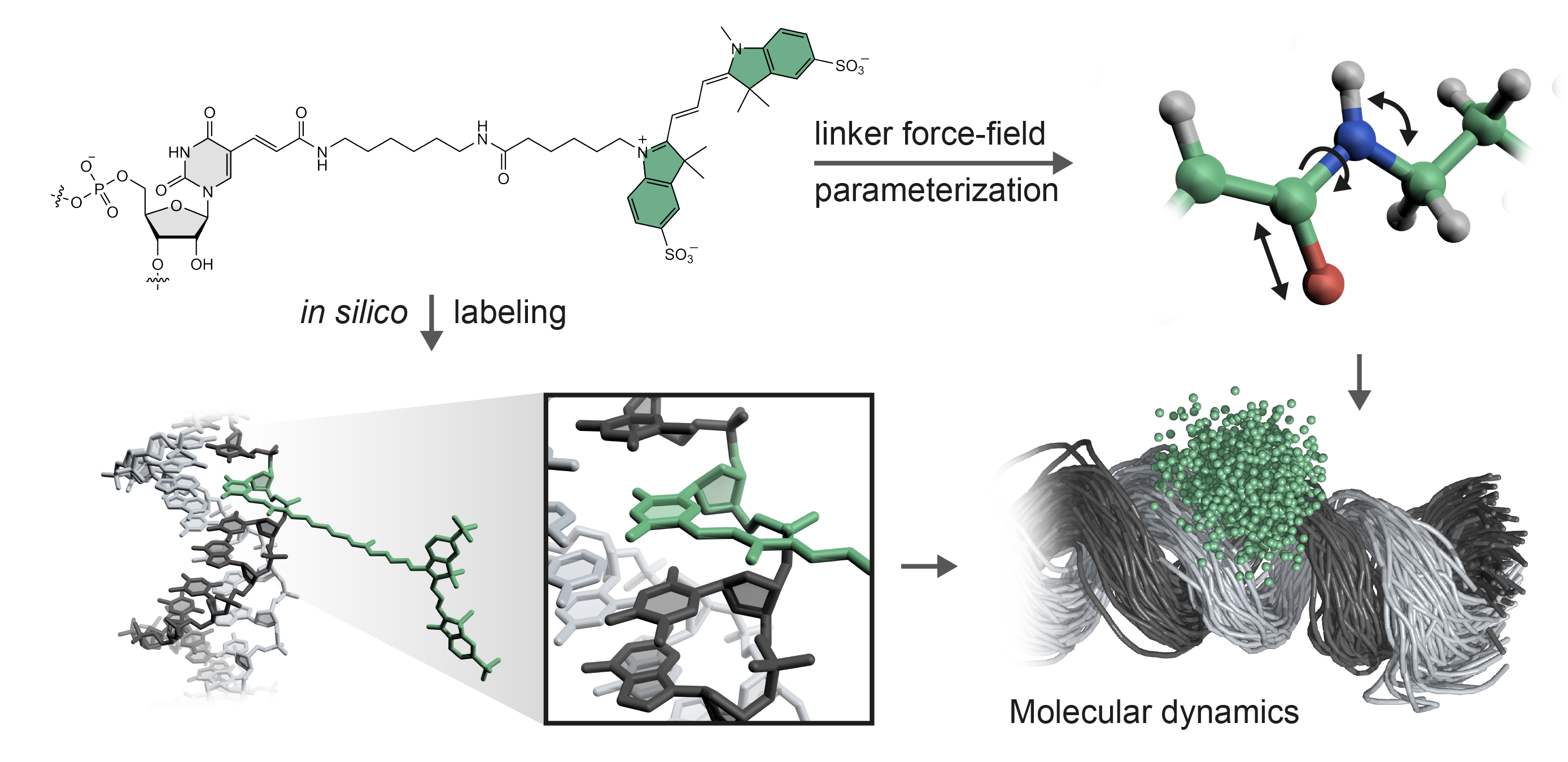
Fig. Schematic of FRETlabel: (i) A fluorescent dye (here Cy3) is coupled to a nucleic acid via a PyMOL plugin. (ii) An existing force field (e.g. AMBERDYES) is patched with parameters for linker fragments to enable MD simulations with explicit fluorophores (dots represent the spatial distribution of the dye).
Installation and Documentation
Follow the instructions for your platform here
FRETlabel and FRETraj
FRETlabel attaches explicit fluorophores on a custom nucleic acid. If you instead like to use an implicit, geometrical dye model that relies on accessible-contact volumes (ACV) then have a look our sister project FRETraj (Steffen, Bioinformatics, 2021)
References
If you use FRETlabel in your work please refer to the following paper:
Additional readings
- T. Graen, M. Hoefling, H. Grubmüller, J. Chem. Theory Comput. 2014, 10, 5505-5512.
- B. Schepers, H. Gohlke, J. Chem. Phys. 2020, 152, 221103.
- R. Shaw, T. Johnston-Wood, B. Ambrose, T. D. Craggs, and J. G. Hill, J. Chem. Theory Comput., 2020, 16, 7817–7824.
- M. Zhao, F.D. Steffen R. Börner, M. Schaffer, R.K.O. Sigel, E. Freisinger, Nucleic Acids Res. 2018, 46, e13.
- F.D. Steffen, R. Börner, E. Freisinger, R.K.O. Sigel, CHIMIA 2019, 73, 257-261.
- F.D. Steffen, R.K.O. Sigel, R. Börner, Bioinformatics 2021, 37, 3953–3955.
1 PyMOL is a trademark of Schrödinger, LLC.
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
Filter files by name, interpreter, ABI, and platform.
If you're not sure about the file name format, learn more about wheel file names.
Copy a direct link to the current filters
File details
Details for the file fretlabel-0.2.1.tar.gz.
File metadata
- Download URL: fretlabel-0.2.1.tar.gz
- Upload date:
- Size: 32.5 kB
- Tags: Source
- Uploaded using Trusted Publishing? Yes
- Uploaded via: twine/6.1.0 CPython/3.12.9
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
e4152d11a2985f5408288a3a0d61735f049591c485ee030303316646585f93e8
|
|
| MD5 |
4c549e01842660efeb142ba4c7722c25
|
|
| BLAKE2b-256 |
84faccbe813a84ef8f49e678f09a2a8634129756dd7847b0fb9ebf64cfad8689
|
Provenance
The following attestation bundles were made for fretlabel-0.2.1.tar.gz:
Publisher:
build.yml on RNA-FRETools/fretlabel
-
Statement:
-
Statement type:
https://in-toto.io/Statement/v1 -
Predicate type:
https://docs.pypi.org/attestations/publish/v1 -
Subject name:
fretlabel-0.2.1.tar.gz -
Subject digest:
e4152d11a2985f5408288a3a0d61735f049591c485ee030303316646585f93e8 - Sigstore transparency entry: 183022040
- Sigstore integration time:
-
Permalink:
RNA-FRETools/fretlabel@041492fc4a272015f9716b239b7c805be2b4afe9 -
Branch / Tag:
refs/tags/0.2.1 - Owner: https://github.com/RNA-FRETools
-
Access:
public
-
Token Issuer:
https://token.actions.githubusercontent.com -
Runner Environment:
github-hosted -
Publication workflow:
build.yml@041492fc4a272015f9716b239b7c805be2b4afe9 -
Trigger Event:
push
-
Statement type:
File details
Details for the file fretlabel-0.2.1-py3-none-any.whl.
File metadata
- Download URL: fretlabel-0.2.1-py3-none-any.whl
- Upload date:
- Size: 30.7 kB
- Tags: Python 3
- Uploaded using Trusted Publishing? Yes
- Uploaded via: twine/6.1.0 CPython/3.12.9
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
5f074a4bc52082df79c91b45012e1c6e9f9df6bcd6b3c5924c4fb0dde4928909
|
|
| MD5 |
63221f5277434ea4d6e2ee586ab21781
|
|
| BLAKE2b-256 |
c7b5b18a0fbf858949f0698fa41cff1e6e6a85c8b849f22ec053004379add65e
|
Provenance
The following attestation bundles were made for fretlabel-0.2.1-py3-none-any.whl:
Publisher:
build.yml on RNA-FRETools/fretlabel
-
Statement:
-
Statement type:
https://in-toto.io/Statement/v1 -
Predicate type:
https://docs.pypi.org/attestations/publish/v1 -
Subject name:
fretlabel-0.2.1-py3-none-any.whl -
Subject digest:
5f074a4bc52082df79c91b45012e1c6e9f9df6bcd6b3c5924c4fb0dde4928909 - Sigstore transparency entry: 183022042
- Sigstore integration time:
-
Permalink:
RNA-FRETools/fretlabel@041492fc4a272015f9716b239b7c805be2b4afe9 -
Branch / Tag:
refs/tags/0.2.1 - Owner: https://github.com/RNA-FRETools
-
Access:
public
-
Token Issuer:
https://token.actions.githubusercontent.com -
Runner Environment:
github-hosted -
Publication workflow:
build.yml@041492fc4a272015f9716b239b7c805be2b4afe9 -
Trigger Event:
push
-
Statement type:

















