No project description provided
Project description
Global-Chem: Collections of common small molecules and their SMILES/SMARTS to support diverse chemical communities
Global Chem is an open-source record collection for common and rare chemical lists using IUPAC as input and SMILES/SMARTS as output. As mostly needed by myself as I search through chemical infinity.
I have found these lists written in history to be useful, they come from a variety of different fields but are aggregated into the most common format of organic chemists (IUPAC) and the common language of the cheminformatician (SMILES) and for pattern matching (SMARTS).

Installation
GlobalChem is going to be distribute via PyPi and as the content store grows we can expand it to other pieces of software making it accessible to all regardless of what you use. Alternatively, you could have a glance at the source code and copy/paste it yourself.
pip install global-chem
Quick Start
Just with no dependencies, intialize the class and there you go! All the common and rare groups of the world at your disposal
To Access Nodes and Visualize the Internal Network:
from global_chem import GlobalChem
gc = GlobalChem()
nodes_list = gc.check_available_nodes()
print (nodes_list)
>>>
'emerging_perfluoro_alkyls', 'montmorillonite_adsorption', 'common_monomer_repeating_units', 'electrophilic_warheads_for_kinases',
gc.build_global_chem_network(print_output=True)
>>>
'global_chem': {
'children': [
'environment',
'miscellaneous',
'organic_synthesis',
'medicinal_chemistry',
'narcotics',
'interstellar_space',
'proteins',
'materials'
],
'name': 'global_chem',
'node_value': <global_chem.global_chem.Node object at 0x10f60eed0>,
'parents': []
},
Fetch Data from Node:
gc = GlobalChem()
gc.build_global_chem_network(print_output=True, debugger=False)
node = gc.get_node('emerging_perfluoroalkyls')
print (node)
>>>
{'perfluorohexanoic acid': 'C(=O)(C(C(C(C(C(F)(F)F)(F)F)(F)F)(F)F)(F)F)O' etc...}
Fetch All Data from Node:
gc = GlobalChem()
print(gc.get_all_smiles())
>>>
['C(=O)(C(C(C(C(C(F)(F)F)(F)F)(F)F)(F)F)(F)F)O', 'C(=O)(C(C(C(C(C(C(F)(F)F)(F)F)(F)F)(F)F)(F)F)(F)F)O' etc...]
Compute Common Score for an IUPAC Name:
gc = GlobalChem()
gc.build_global_chem_network(print_output=False, debugger=False)
gc.compute_common_score('benzene', verbose=True)
To Create Your Own Chemical Graph Network And Check the Values
from global_chem import GlobalChem
gc = GlobalChem(verbose=False)
gc.initiate_network()
gc.add_node('global_chem', 'common_monomer_repeating_units')
gc.add_node('common_monomer_repeating_units','electrophilic_warheads_for_kinases')
values = gc.get_node_smiles('common_monomer_repeating_units')
print (values)
>>>
'3′-bromo-2-chloro[1,1′:4′,1′′-terphenyl]-4,4′′': 'ClC1=CC=CC=C1C2=CC=C(C3=CC=CC=C3)C(Br)=C2'
values = gc.get_node_smarts('electrophilic_warheads_for_kinases')
>>>
'propane-1,3-diyl': '[#6]-[#6]-[#6]', 'methylmethylene': '[#6H]-[#6]',
Variables List
| Chemical List | # of Entries | References |
|---|---|---|
| Amino Acids | 20 | Common Knowledge |
| Essential Vitamins | 13 | Common Knowledge |
| Common Organic Solvents | 42 | Fulmer, Gregory R., et al. “NMR Chemical Shifts of Trace Impurities: Common Laboratory Solvents, Organics, and Gases in Deuterated Solvents Relevant to the Organometallic Chemist.”Organometallics, vol. 29, no. 9, May 2010, pp. 2176–79. |
| Open Smiles | 94 | OpenSMILES Home Page. http://opensmiles.org/. |
| IUPAC Blue Book (CRC Handbook) 2003 | 333 | Chemical Rubber Company. CRC Handbook of Chemistry and Physics: A Ready-Reference Book of Chemical and Physical Data Edited by David R. Lide, 85. ed, CRC Press, 2004. |
| Rings in Drugs | 92 | Taylor, Richard D., et al. “Rings in Drugs.” Journal of Medicinal Chemistry, vol. 57, no. 14, July 2014, pp. 5845–59. ACS Publications, https://doi.org/10.1021/jm4017625. |
| Phase 2 Hetereocyclic Rings | 19 | Broughton, Howard B., and Ian A. Watson. “Selection of Heterocycles for Drug Design.” Journal of Molecular Graphics & Modelling, vol. 23, no. 1, Sept. 2004, pp. 51–58. PubMed, https://doi.org/10.1016/j.jmgm.2004.03.016. |
| Privileged Scaffolds | 47 | Welsch, Matthew E., et al. “Privileged Scaffolds for Library Design and Drug Discovery.” Current Opinion in Chemical Biology , vol. 14, no. 3, June 2010, pp. 347–61.PubMed, https://doi.org/10.1016/j.cbpa.2010.02.018. |
| Common Warheads | 29 | Gehringer, Matthias, and Stefan A. Laufer. “Emerging and Re-Emerging Warheads for Targeted Covalent Inhibitors: Applications in Medicinal Chemistry and Chemical Biology.”Journal of Medicinal Chemistry , vol. 62, no. 12, June 2019, pp. 5673–724. ACS Publications, https://doi.org/10.1021/acs.jmedchem.8b01153. |
| Common Polymer Repeating Units | 78 | Hiorns, R. C., et al. “A brief guide to polymer nomenclature (IUPAC Technical Report).”Pure and Applied Chemistry , vol. 84, no. 10, Oct. 2012, pp. 2167–69., https://doi.org/10.1351/PAC-REP-12-03-05. |
| Common R Group Replacements | 499 | Takeuchi, Kosuke, et al. “R-Group Replacement Database for Medicinal Chemistry.” Future Science OA , vol. 7, no. 8, Sept. 2021, p. FSO742. future-science.com (Atypon) , https://doi.org/10.2144/fsoa-2021-0062. |
| Electrophillic Warheads for Kinases | 24 | Petri, László, et al. “An Electrophilic Warhead Library for Mapping the Reactivity and Accessibility of Tractable Cysteines in Protein Kinases.” European Journal of Medicinal Chemistry, vol. 207, Dec. 2020, p. 112836. PubMed, https://doi.org/10.1016/j.ejmech.2020.112836. |
| Privileged Scaffolds for Kinases | 29 | Hu, Huabin, et al. “Systematic Comparison of Competitive and Allosteric Kinase Inhibitors Reveals Common Structural Characteristics.” European Journal of Medicinal Chemistry, vol. 214, Mar. 2021, p. 113206. ScienceDirect, https://doi.org/10.1016/j.ejmech.2021.113206. |
| BRaf Inhibitors | 54 | Agianian, Bogos, and Evripidis Gavathiotis. “Current Insights of BRAF Inhibitors in Cancer.” Journal of Medicinal Chemistry, vol. 61, no. 14, July 2018, pp. 5775–93. ACS Publications, https://doi.org/10.1021/acs.jmedchem.7b01306. |
| Common Amino Acid Protecting Groups | 346 | Isidro-Llobet, Albert, et al. “Amino Acid-Protecting Groups.” Chemical Reviews, vol. 109, no. 6, June 2009, pp. 2455–504. DOI.org (Crossref), https://doi.org/10.1021/cr800323s. |
| Emerging Perfluoroalkyls | 27 | Pelch, Katherine E., et al. “PFAS Health Effects Database: Protocol for a Systematic Evidence Map.” Environment International, vol. 130, Sept. 2019, p. 104851. ScienceDirect, https://doi.org/10.1016/j.envint.2019.05.045. |
| Chemicals For Clay Adsorption | 33 | Orr, Asuka A., et al. “Combining Experimental Isotherms, Minimalistic Simulations, and a Model to Understand and Predict Chemical Adsorption onto Montmorillonite Clays.” ACS Omega, vol. 6, no. 22, June 2021, pp. 14090–103. PubMed, https://doi.org/10.1021/acsomega.1c00481. |
| Cannabinoids | 63 | Turner, Carlton E., et al. “Constituents of Cannabis Sativa L. XVII. A Review of the Natural Constituents.” Journal of Natural Products, vol. 43, no. 2, Mar. 1980, pp. 169–234. ACS Publications, https://doi.org/10.1021/np50008a001. |
| Schedule 1 United States Narcotics | 240 | ECFR :: 21 CFR Part 1308 - Schedules. |
| Schedule 2 United States Narcotics | 60 | ECFR :: 21 CFR Part 1308 - Schedules. |
| Schedule 3 United States Narcotics | 22 | ECFR :: 21 CFR Part 1308 - Schedules. |
| Schedule 4 United States Narcotics | 77 | ECFR :: 21 CFR Part 1308 - Schedules. |
| Schedule 5 United States Narcotics | 8 | ECFR :: 21 CFR Part 1308 - Schedules. |
| Common Regex Patterns | 1 |
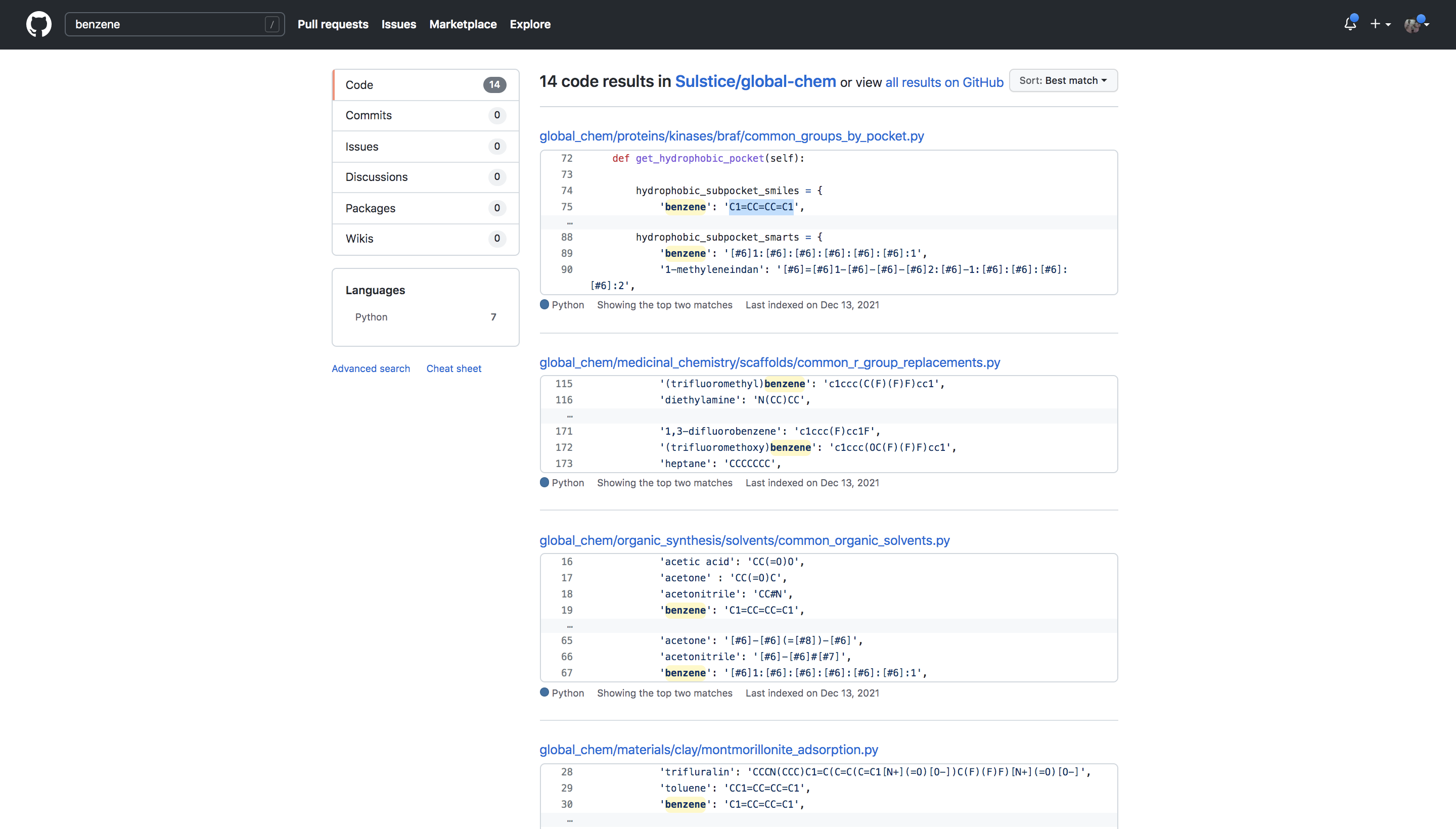
GlobalChem, initially, is one class object with a series of Nodes that are act as objects for any common chemical lists. The chemical lists can be accessed as nodes and the user can construct their own node trees for the lists.
Also since these lists of commonality are stored on github it is easily searchable and tied directly to the paper for any bypasser.
 >>>>>>> 713c3366fce5a5a3afb0b0c478f1f50048cb07c2
>>>>>>> 713c3366fce5a5a3afb0b0c478f1f50048cb07c2
Genesis
GlobalChem was created because I noticed I was using the same variable across multiple scripts and figure it would be useful for folk to have.
- Lead Developer Suliman sharif
- Artwork Elena Chow
Citation
It's on it's way
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.






















