GO enrichment with python -- pandas meets networkx
Project description
goenrich
Convenient GO enrichments from python. For use in python projects.
Builds the GO-ontology graph
Propagates GO-annotations up the graph
Performs enrichment test for all categories
Performs multiple testing correction
Allows for export to pandas for processing and graphviz for visualization
Supported ids: Uniport ACC, Entrez GeneID
Installation
Install package from pypi and download ontology
and needed annotations.
pip install goenrich
mkdir db
# Ontology
wget http://purl.obolibrary.org/obo/go/go-basic.obo -O db/go-basic.obo
# UniprotACC
wget http://geneontology.org/gene-associations/gene_association.goa_ref_human.gz -O db/gene_association.goa_ref_human.gz
# Entrez GeneID
wget ftp://ftp.ncbi.nlm.nih.gov/gene/DATA/gene2go.gz -O db/gene2go.gzRun GO enrichment
import goenrich
# build the ontology
G = goenrich.obo.graph('db/go-basic.obo')
# use all entrez geneid associations form gene2go as background
# use goenrich.read.goa('db/gene_association.goa_ref_human.gz') for uniprot
background = goenrich.read.gene2go('db/gene2go.gz')
goenrich.enrich.set_background(G, background, 'GeneID', 'GO_ID')
# extract some list of entries as example query
query = set(background['GeneID'].unique()[:20])
# run analysis and obtain results
result = goenrich.enrich.analyze(G, query)
# for additional export to graphviz just specify the gvfile argument
# the show argument keeps the graph reasonably small
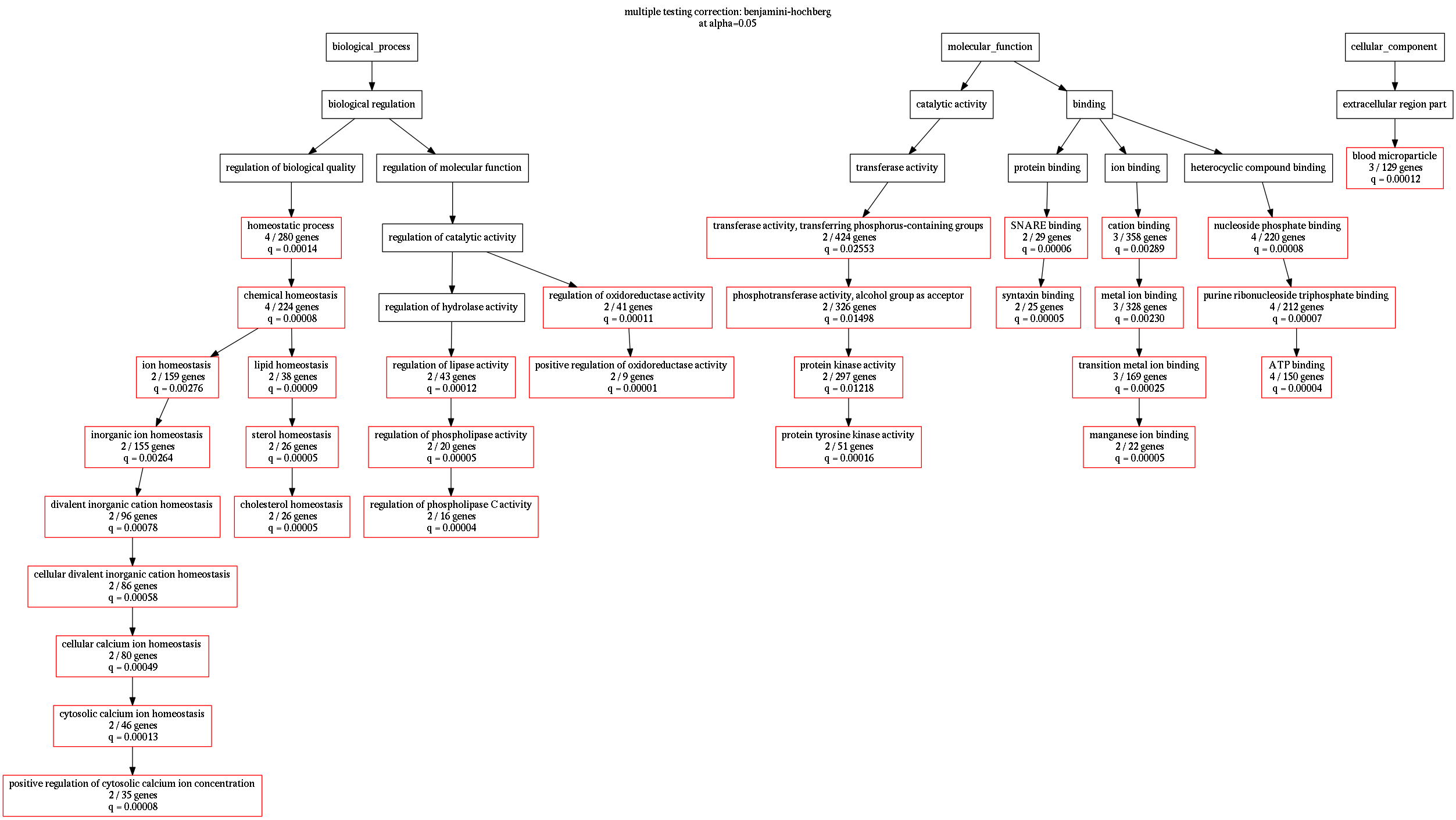
result = goenrich.enrich.analyze(G, query, gvfile='example.dot', show='top20')Generate png image using graphviz
dot -Tpng example.dot > example.pngParameters
Parameters can all be passed to enrich.analyze as shown below
go_options = {
'multiple-testing-correction' : 'bonferroni',
'alpha' : 0.05,
'node_filter' : lambda x : x.get('significant', False)
}
goenrich.enrich.analyze(G, query, **go_options)
# export results to graphviz
goenrich.enrich.analyze(G, query, gvfile='example.dot', **go_options)Here is an overview over the available parmeters
enrich.analyze: node_filter = lambda node : 'p' in node show = 'top20' # works for any 'topNUM' enrich.calculate_pvalues: min_category_size = 3 max_category_size = 500 min_hit_size = 2 enrich.multiple_testing_correction: alpha = 0.05 method = ['benjamin-hochberg', 'bonferroni'] export.to_frame: node_filter = lambda node: True export.to_graphviz: graph_label = None # if None it is replaced by multiple testing info
Licence
This work is licenced under the MIT licence
Contributions are welcome!
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
goenrich-1.1.0.tar.gz
(7.1 kB
view hashes)