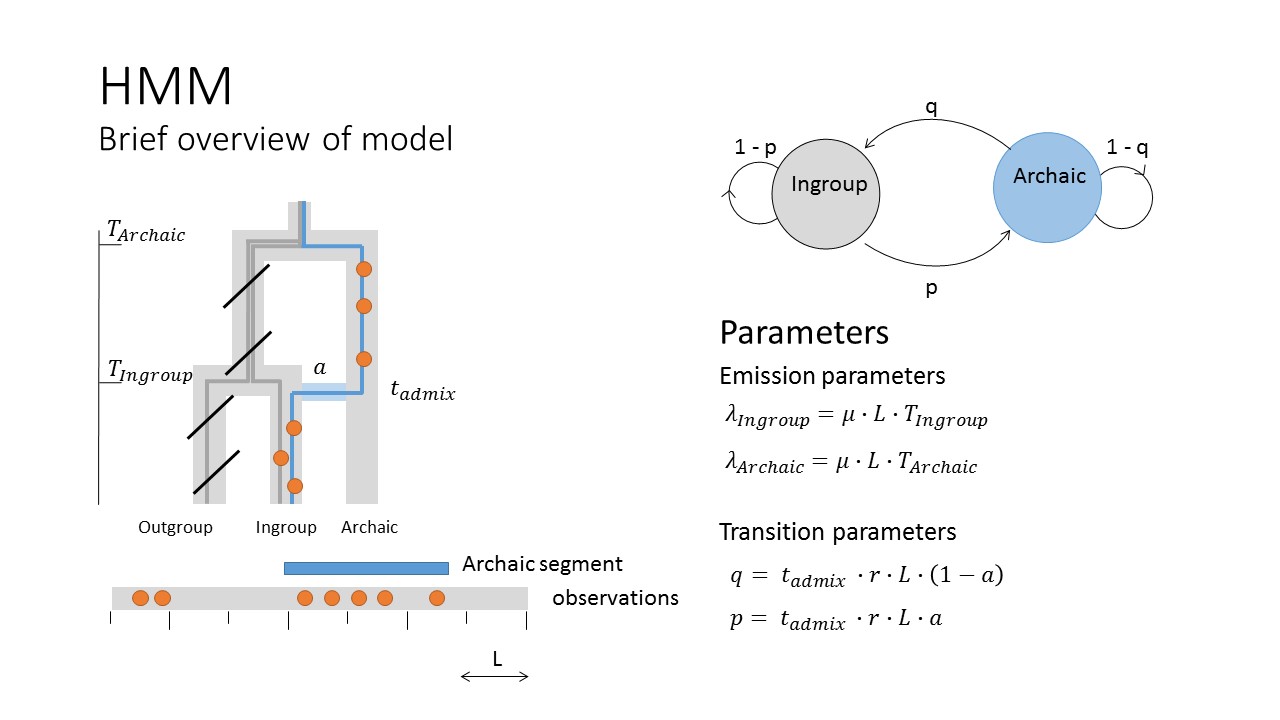
Find introgressed segments
Project description
Introgression detection
Contact
https://sites.google.com/view/laurits-skov
Helpful files
The outgroup files, mutation rate files, reference genomes, ancestral alleles and callability files and ancestral allele files are now premade!
https://doi.org/10.5281/zenodo.11212339 (hg19 and hg38)
Using these files I have already called archaic segments in 1000 genomes and HDGP datasets (hg38 reference coordinate system)
https://doi.org/10.5281/zenodo.14136628 (archaic introgression callsets for HGDP and 1000genomes in hg38)
VCF file containing 4 high coverage archaic genomes (Altai, Vindija and Chagyrskaya Neanderthals and Denisovan) here:
https://zenodo.org/records/7246376 (hg19)
https://zenodo.org/records/13368126 (hg38)
If you are working with archaic introgression into present-day humans of non-African ancestry you can use these files and skip the following steps: Find derived variants in outgroup and Estimate local mutation rate.
These are the scripts needed to infere archaic introgression in modern populations using an unadmixed outgroup.
Installation
Run the following command to install:
pip install hmmix
If you want to work with bcf/vcf files you should also install vcftools and bcftools. You can either use conda or visit their websites.
conda install -c bioconda vcftools bcftools
The way the model works is by removing variation found in an outgroup population and then using the remaining variants to group the genome into regions of different variant density. If the model works well we would expect that introgressed regions have higher variant density than non-introgressed - because they have spend more time accumulation variation that is not found in the outgroup.
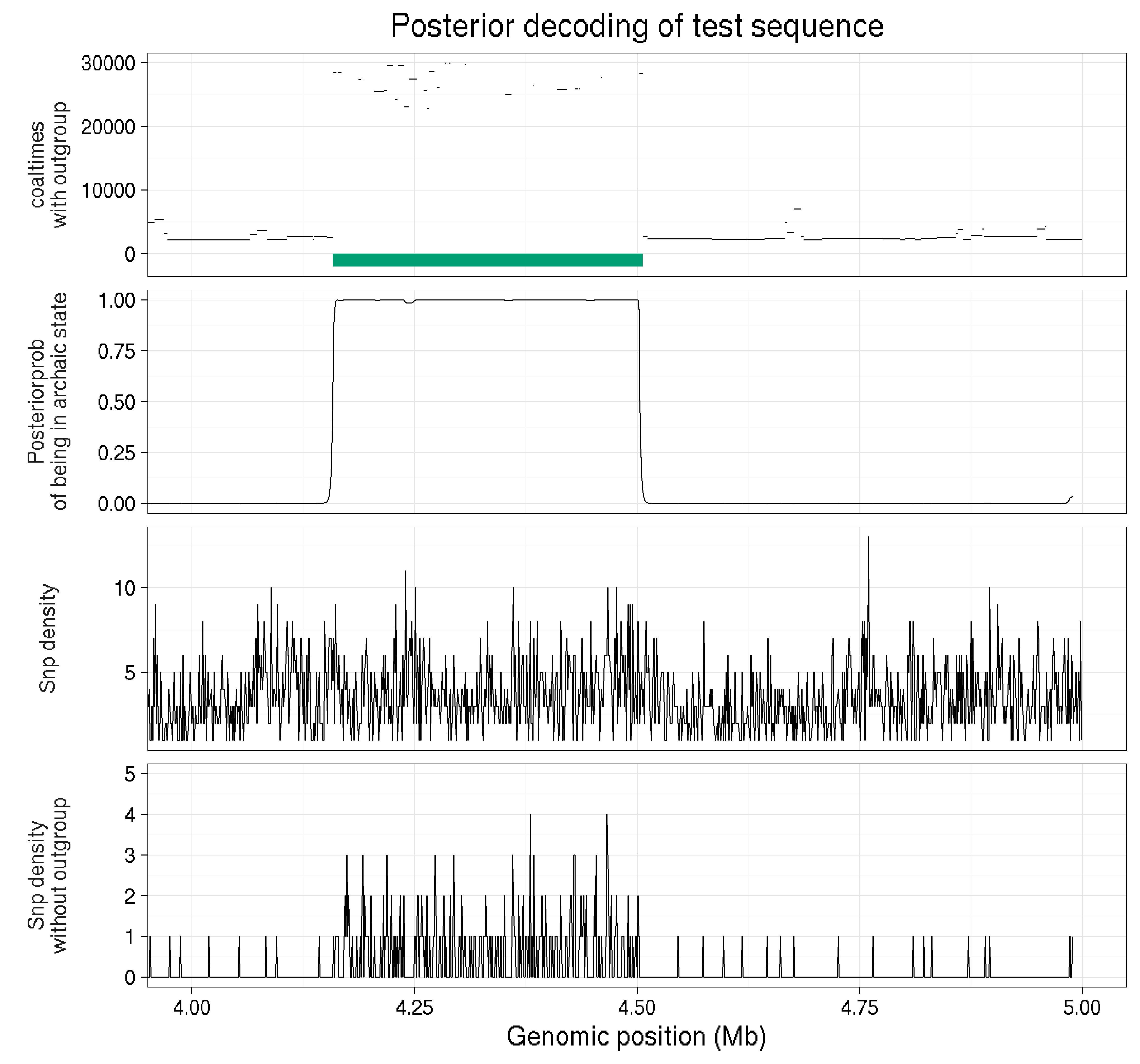
An example on simulated data is provided below:
In this example we zoom in on 1 Mb of simulated data for a haploid genome. The top panel shows the coalescence times with the outgroup across the region and the green segment is an archaic introgressed segment. Notice how much more deeper the coalescence time with the outgroup is. The second panel shows that probability of being in the archaic state. We can see that the probability is much higher in the archaic segment, demonstrating that in this toy example the model is working like we would hope. The next panel is the snp density if you dont remove all snps found in the outgroup. By looking at this one cant tell where the archaic segments begins and ends, or even if there is one. The bottom panel is the snp density when all variation in the outgroup is removed. Notice that now it is much clearer where the archaic segment begins and ends!
The method is now published in PlosGenetics and can be found here: Detecting archaic introgression using an unadmixed outgroup This paper is describing and evaluating the method.
Usage
Script for identifying introgressed archaic segments
> Turorial:
hmmix make_test_data
hmmix train -obs=obs.txt -weights=weights.bed -mutrates=mutrates.bed -param=Initialguesses.json -out=trained.json
hmmix decode -obs=obs.txt -weights=weights.bed -mutrates=mutrates.bed -param=trained.json
Different modes (you can also see the options for each by writing hmmix make_test_data -h):
> make_test_data
-windows Number of Kb windows to create (defaults to 50,000 per chromosome)
-chromosomes Number of chromosomes to simulate (defaults to 2)
-nooutfiles Don't create obs.txt, mutrates.bed, weights.bed, Initialguesses.json, simulated_segments.txt (defaults to yes)
-param markov parameters file (default is human/neanderthal like parameters)
> mutation_rate
-outgroup [required] path to variants found in outgroup
-out outputfile (defaults to mutationrate.bed)
-weights file with callability (defaults to all positions being called)
-window_size size of bins (defaults to 1 Mb)
> create_outgroup
-ind [required] ingroup/outgrop list (json file) or comma-separated list e.g. ind1,ind2
-vcf [required] path to list of comma-separated vcf/bcf file(s) or wildcard characters e.g. chr*.bcf
-weights file with callability (defaults to all positions being called)
-out outputfile (defaults to stdout)
-ancestral fasta file with ancestral information - comma-separated list or wildcards like vcf argument (default none)
-refgenome fasta file with reference genome - comma-separated list or wildcards like vcf argument (default none)
> create_ingroup
-ind [required] ingroup/outgrop list (json file) or comma-separated list e.g. ind1,ind2
-vcf [required] path to list of comma-separated vcf/bcf file(s) or wildcard characters e.g. chr*.bcf
-outgroup [required] path to variant found in outgroup
-weights file with callability (defaults to all positions being called)
-out outputfile prefix (default is a file named obs.<ind>.txt where ind is the name of individual in ingroup/outgrop list)
-ancestral fasta file with ancestral information - comma-separated list or wildcards like vcf argument (default none)
> train
-obs [required] file with observation data
-chrom Subset to chromosome or comma separated list of chromosomes e.g chr1 or chr1,chr2,chr3 (default is use all chromosomes)
-weights file with callability (defaults to all positions being called)
-mutrates file with mutation rates (default is mutation rate is uniform)
-param markov parameters file (default is human/neanderthal like parameters)
-out outputfile (default is a file named trained.json)
-window_size size of bins (default is 1000 bp)
-haploid Change from using diploid data to haploid data (default is diploid)
> decode
-obs [required] file with observation data
-chrom Subset to chromosome or comma separated list of chromosomes e.g chr1 or chr1,chr2,chr3 (default is use all chromosomes)
-weights file with callability (defaults to all positions being called)
-mutrates file with mutation rates (default is mutation rate is uniform)
-param markov parameters file (default is human/neanderthal like parameters)
-out outputfile prefix <out>.hap1.txt and <out>.hap2.txt if -haploid option is used or <out>.diploid.txt (default is stdout)
-window_size size of bins (default is 1000 bp)
-haploid Change from using diploid data to haploid data (default is diploid)
-admixpop Annotate using vcffile with admixing population (default is none)
-extrainfo Add variant position for each SNP (default is off)
-viterbi decode using the viterbi algorithm (default is posterior decoding)
> inhomogeneous
-obs [required] file with observation data
-chrom Subset to chromosome or comma separated list of chromosomes e.g chr1 or chr1,chr2,chr3 (default is use all chromosomes)
-weights file with callability (defaults to all positions being called)
-mutrates file with mutation rates (default is mutation rate is uniform)
-param markov parameters file (default is human/neanderthal like parameters)
-out outputfile prefix <out>.hap1_sim(0-n).txt and <out>.hap2_sim(0-n).txt if -haploid option is used or <out>.diploid_(0-n).txt (default is stdout)
-window_size size of bins (default is 1000 bp)
-haploid Change from using diploid data to haploid data (default is diploid)
-samples Number of simulated paths for the inhomogeneous markov chain (default is 100)
-admixpop Annotate using vcffile with admixing population (default is none)
-extrainfo Add variant position for each SNP (default is off)
Quick tutorial
Here is how we can simulate test data using hmmix. Lets make some test data and start using the program.
> hmmix make_test_data
> creating 2 chromosomes each with 50000 kb of test data with the following parameters..
> hmm parameters file: None
> state_names = ['Human', 'Archaic']
> starting_probabilities = [0.98, 0.02]
> transitions = [[1.0, 0.0], [0.02, 0.98]]
> emissions = [0.04, 0.4]
> Seed is 42
This will generate 5 files, obs.txt, weights.bed, mutrates.bed, simulated_segments.txt and Initialguesses.json. obs.txt. These are the mutations that are left after removing variants which are found in the outgroup.
chrom pos ancestral_base genotype
chr1 17102 C CT
chr1 34435 C CT
chr1 69860 T TA
chr1 122270 C CA
chr1 181106 G GC
chr1 218071 A AC
chr1 220700 T TG
chr1 231020 A AG
chr1 235614 T TG
weights.bed. This is the parts of the genome that we can accurately map to - in this case we have simulated the data and can accurately access the entire genome.
chr1 0 50000000
chr2 0 50000000
mutrates.bed. This is the normalized mutation rate across the genome.
chr1 0 50000000 1
chr2 0 50000000 1
Initialguesses.json. This is our initial guesses when training the model - note these are different from those we simulated from.
{
"state_names": ["Human","Archaic"],
"starting_probabilities": [0.5,0.5],
"transitions": [[0.99,0.01],[0.02,0.98]],
"emissions": [0.03,0.3]
}
The simulated_segments.txt contains the simulated states which generated the data (you can compare this to the decoded results later and see that it matches).
chrom start end length state
chr1 0 22980000 22980000 Human
chr1 22980000 23071000 91000 Archaic
chr1 23071000 43905000 20834000 Human
chr1 43905000 43911000 6000 Archaic
chr1 43911000 47419000 3508000 Human
chr1 47419000 47443000 24000 Archaic
chr1 47443000 50000000 2557000 Human
chr2 0 16378000 16378000 Human
chr2 16378000 16492000 114000 Archaic
chr2 16492000 19478000 2986000 Human
chr2 19478000 19512000 34000 Archaic
chr2 19512000 37728000 18216000 Human
chr2 37728000 37751000 23000 Archaic
chr2 37751000 46777000 9026000 Human
chr2 46777000 46791000 14000 Archaic
chr2 46791000 50000000 3209000 Human
We can find the best fitting parameters using BaumWelsch training. Here is how you use it: - note you can try to ommit the weights and mutrates arguments. Since this is simulated data the mutation is constant across the genome and we can asses the entire genome. Also notice how the parameters approach the parameters the data was generated from (jubii).
> hmmix train -obs=obs.txt -weights=weights.bed -mutrates=mutrates.bed -param=Initialguesses.json -out=trained.json
----------------------------------------
> state_names = ['Human', 'Archaic']
> starting_probabilities = [0.5, 0.5]
> transitions = [[0.99, 0.01], [0.02, 0.98]]
> emissions = [0.03, 0.3]
> chromosomes to use: All
> number of windows: 99982. Number of snps = 4116
> total callability: 99982000 bp (100.0 %)
> average mutation rate per bin: 1.0
> Output is trained.json
> Window size is 1000 bp
> Haploid False
----------------------------------------
iteration loglikelihood start1 start2 emis1 emis2 trans1_1 trans2_2
0 -17904.2589 0.5 0.5 0.03 0.3 0.99 0.98
1 -17259.0236 0.96 0.04 0.0346 0.2009 0.9968 0.9217
2 -17243.7038 0.969 0.031 0.0365 0.1861 0.9971 0.9105
...
29 -17195.4135 0.997 0.003 0.0401 0.4477 0.9999 0.9803
30 -17195.4098 0.997 0.003 0.0401 0.4482 0.9999 0.9806
31 -17195.409 0.997 0.003 0.0401 0.4485 0.9999 0.9808
# run without mutrate and weights (only do this for simulated data)
> hmmix train -obs=obs.txt -param=Initialguesses.json -out=trained.json
We can now decode the data with the best parameters that maximize the likelihood and find the archaic segments. Please note that even thought we simulated 50 Mb of sequence the last window in the decoded output is at window 49,985,000 for chromosome 1 and 49,997,000 for chromosome 2. This is because hmmix uses the position of the last SNP to determine the length of the chromosome. The last SNP on chromosome 1 is 49,984,119 and the last SNP on chromosome 2 is 49,996,253.
> hmmix decode -obs=obs.txt -weights=weights.bed -mutrates=mutrates.bed -param=trained.json
----------------------------------------
> state_names = ['Human', 'Archaic']
> starting_probabilities = [0.997, 0.003]
> transitions = [[1.0, 0.0], [0.019, 0.981]]
> emissions = [0.04, 0.448]
> chromosomes to use: All
> number of windows: 99982. Number of snps = 4116
> total callability: 99982000 bp (100.0 %)
> average mutation rate per bin: 1.0
> Output prefix is /dev/stdout
> Window size is 1000 bp
> Haploid False
> Decode with posterior decoding
----------------------------------------
chrom start end length state mean_prob snps
chr1 0 22979000 22979000 Human 0.99989 903
chr1 22979000 23066000 87000 Archaic 0.96368 32
chr1 23066000 47418000 24352000 Human 0.99975 935
chr1 47418000 47443000 25000 Archaic 0.88224 10
chr1 47443000 49985000 2542000 Human 0.99933 96
chr2 0 16381000 16381000 Human 0.99981 653
chr2 16381000 16492000 111000 Archaic 0.99166 60
chr2 16492000 19478000 2986000 Human 0.99883 134
chr2 19478000 19512000 34000 Archaic 0.96454 18
chr2 19512000 49997000 30485000 Human 0.99981 1275
Example with 1000 genomes data
The whole pipeline we will run looks like this. In the following section we will go through all the steps on the way
NOTE: The outgroup files, mutation rate files, reference genomes, ancestral alleles and callability files and ancestral allele files are now premade! They can be downloaded in hg38 and hg19 here: https://doi.org/10.5281/zenodo.11212339
But keep reading along if you want to know HOW the files were generated! Another important thing to note is that hmmix is relying on VCFtools which only support VCF files up to format V4.2 - so if you have VCFfiles in version 4.3 you will need to change this in your header!
hmmix create_outgroup -ind=individuals.json -vcf=*.bcf -weights=strickmask.bed -out=outgroup.txt -ancestral=hg19_ancestral/homo_sapiens_ancestor_*.fa -refgenome=hg19_refgenome/*fa
hmmix mutation_rate -outgroup=outgroup.txt -weights=strickmask.bed -window_size=1000000 -out mutationrate.bed
hmmix create_ingroup -ind=individuals.json -vcf=*.bcf -weights=strickmask.bed -out=obs -outgroup=outgroup.txt -ancestral=hg19_ancestral/homo_sapiens_ancestor_*.fa
hmmix train -obs=obs.HG00096.txt -weights=strickmask.bed -mutrates=mutationrate.bed -out=trained.HG00096.json
hmmix decode -obs=obs.HG00096.txt -weights=strickmask.bed -mutrates=mutationrate.bed -param=trained.HG00096.json
Getting data
I thought it would be nice to have an entire reproduceble example of how to use this model. From a common starting point such as a VCF file (well a BCF file in this case) to the final output. The reason for using BCF files is because it is MUCH faster to extract data for each individual. You can convert a vcf file to a bcf file like this:
bcftools view file.vcf -l 1 -O b > file.bcf
bcftools index file.bcf
In this example I will analyse an individual (HG00096) from the 1000 genomes project phase 3. All analysis are run on my lenovo thinkpad (8th gen) computer so it should run on yours too!
First we will need to know which 1) bases can be called in the genome and 2) which variants are found in the outgroup. So let's start out by downloading the files from the following directories. To download callability regions, ancestral alleles information, ingroup outgroup information call this command:
# bcffiles (hg19)
ftp.1000genomes.ebi.ac.uk/vol1/ftp/release/20130502/supporting/bcf_files/
# callability (remember to remove chr in the beginning of each line to make it compatible with hg19 e.g. chr1 > 1)
ftp.1000genomes.ebi.ac.uk/vol1/ftp/release/20130502/supporting/accessible_genome_masks/20141020.strict_mask.whole_genome.bed
sed 's/^chr\|%$//g' 20141020.strict_mask.whole_genome.bed | awk '{print $1"\t"$2"\t"$3}' > strickmask.bed
# outgroup information
ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/release/20130502/integrated_call_samples_v3.20130502.ALL.panel
# Ancestral information
ftp://ftp.ensembl.org/pub/release-74/fasta/ancestral_alleles/hg19_ancestral.tar.bz2
# Reference genome
wget 'ftp://hgdownload.soe.ucsc.edu/goldenPath/hg19/bigZips/chromFa.tar.gz' -O chromFa.tar.gz
# Archaic variants (Altai, Vindija, Chagyrskaya and Denisova in hg19)
https://zenodo.org/records/7246376
For this example we will use all individuals from 'YRI','MSL' and 'ESN' as outgroup individuals. While we will only be decoding HG00096 in this example you can add as many individuals as you want to the ingroup.
{
"ingroup": [
"HG00096",
"HG00097"
],
"outgroup": [
"HG02922",
"HG02923",
...
"HG02944",
"HG02946"]
}
Finding snps which are derived in the outgroup
First we need to find a set of variants found in the outgroup. We can use the wildcard character to loop through all bcf files. It is best if you have files with the ancestral alleles (in FASTA format) and the reference genome (in FASTA format) but the program will run without.
Something to note is that if you use an outgroup vcffile (like 1000 genomes) and an ingroup vcf file from a different dataset (like SGDP) there is an edge case which could occur. There could be recurrent mutations where every individual in 1000 genome has the derived variant and one individual in SGDP where the derived variant has mutated back to the ancestral allele. This means that this position will not be present in the outgroup file. However if a recurrent mutation occurs it will look like multiple individuals in the ingroup file have the mutation. This does not happen often but that is why I recommend having files with the ancestral allele and reference genome information.
# Recommended usage (if you want to remove sites which are fixed derived in your outgroup/ingroup). This is the file from zenodo.
(took two hours) > hmmix create_outgroup -ind=individuals.json -vcf=*.bcf -weights=strickmask.bed -out=outgroup.txt -ancestral=hg19_ancestral/homo_sapiens_ancestor_*.fa -refgenome=hg19_refgenome/*fa
----------------------------------------
> Outgroup individuals: 292
> Using vcf and ancestral files
vcffile: chr1.bcf ancestralfile: hg19_ancestral/homo_sapiens_ancestor_1.fa reffile: hg19_refgenome/chr1.fa
vcffile: chr2.bcf ancestralfile: hg19_ancestral/homo_sapiens_ancestor_2.fa reffile: hg19_refgenome/chr2.fa
vcffile: chr3.bcf ancestralfile: hg19_ancestral/homo_sapiens_ancestor_3.fa reffile: hg19_refgenome/chr3.fa
vcffile: chr4.bcf ancestralfile: hg19_ancestral/homo_sapiens_ancestor_4.fa reffile: hg19_refgenome/chr4.fa
vcffile: chr5.bcf ancestralfile: hg19_ancestral/homo_sapiens_ancestor_5.fa reffile: hg19_refgenome/chr5.fa
vcffile: chr6.bcf ancestralfile: hg19_ancestral/homo_sapiens_ancestor_6.fa reffile: hg19_refgenome/chr6.fa
vcffile: chr7.bcf ancestralfile: hg19_ancestral/homo_sapiens_ancestor_7.fa reffile: hg19_refgenome/chr7.fa
vcffile: chr8.bcf ancestralfile: hg19_ancestral/homo_sapiens_ancestor_8.fa reffile: hg19_refgenome/chr8.fa
vcffile: chr9.bcf ancestralfile: hg19_ancestral/homo_sapiens_ancestor_9.fa reffile: hg19_refgenome/chr9.fa
vcffile: chr10.bcf ancestralfile: hg19_ancestral/homo_sapiens_ancestor_10.fa reffile: hg19_refgenome/chr10.fa
vcffile: chr11.bcf ancestralfile: hg19_ancestral/homo_sapiens_ancestor_11.fa reffile: hg19_refgenome/chr11.fa
vcffile: chr12.bcf ancestralfile: hg19_ancestral/homo_sapiens_ancestor_12.fa reffile: hg19_refgenome/chr12.fa
vcffile: chr13.bcf ancestralfile: hg19_ancestral/homo_sapiens_ancestor_13.fa reffile: hg19_refgenome/chr13.fa
vcffile: chr14.bcf ancestralfile: hg19_ancestral/homo_sapiens_ancestor_14.fa reffile: hg19_refgenome/chr14.fa
vcffile: chr15.bcf ancestralfile: hg19_ancestral/homo_sapiens_ancestor_15.fa reffile: hg19_refgenome/chr15.fa
vcffile: chr16.bcf ancestralfile: hg19_ancestral/homo_sapiens_ancestor_16.fa reffile: hg19_refgenome/chr16.fa
vcffile: chr17.bcf ancestralfile: hg19_ancestral/homo_sapiens_ancestor_17.fa reffile: hg19_refgenome/chr17.fa
vcffile: chr18.bcf ancestralfile: hg19_ancestral/homo_sapiens_ancestor_18.fa reffile: hg19_refgenome/chr18.fa
vcffile: chr19.bcf ancestralfile: hg19_ancestral/homo_sapiens_ancestor_19.fa reffile: hg19_refgenome/chr19.fa
vcffile: chr20.bcf ancestralfile: hg19_ancestral/homo_sapiens_ancestor_20.fa reffile: hg19_refgenome/chr20.fa
vcffile: chr21.bcf ancestralfile: hg19_ancestral/homo_sapiens_ancestor_21.fa reffile: hg19_refgenome/chr21.fa
vcffile: chr22.bcf ancestralfile: hg19_ancestral/homo_sapiens_ancestor_22.fa reffile: hg19_refgenome/chr22.fa
vcffile: chrX.bcf ancestralfile: hg19_ancestral/homo_sapiens_ancestor_X.fa reffile: hg19_refgenome/chrX.fa
> Callability file: strickmask.bed
> Writing output to: outgroup.txt
----------------------------------------
Here it is important to check that hmmix matches up the reference, ancestral and vcffiles correctly e.g. chr1.bcf should fit with hg19_ancestral/homo_sapiens_ancestor_1.fa and hg19_refgenome/chr1.fa for instance. If you see an issue here its better to give the files as commaseparated values.
hmmix create_outgroup -ind=individuals.json \
-vcf=chr1.bcf,chr2.bcf,chr3.bcf,chr4.bcf,chr5.bcf,chr6.bcf,chr7.bcf,chr8.bcf,chr9.bcf,chr10.bcf,chr11.bcf,chr12.bcf,chr13.bcf,chr14.bcf,chr15.bcf,chr16.bcf,chr17.bcf,chr18.bcf,chr19.bcf,chr20.bcf,chr21.bcf,chr22.bcf,chrX.bcf \
-ancestral=hg19_ancestral/homo_sapiens_ancestor_1.fa,hg19_ancestral/homo_sapiens_ancestor_2.fa,hg19_ancestral/homo_sapiens_ancestor_3.fa,hg19_ancestral/homo_sapiens_ancestor_4.fa,hg19_ancestral/homo_sapiens_ancestor_5.fa,hg19_ancestral/homo_sapiens_ancestor_6.fa,hg19_ancestral/homo_sapiens_ancestor_7.fa,hg19_ancestral/homo_sapiens_ancestor_8.fa,hg19_ancestral/homo_sapiens_ancestor_9.fa,hg19_ancestral/homo_sapiens_ancestor_10.fa,hg19_ancestral/homo_sapiens_ancestor_11.fa,hg19_ancestral/homo_sapiens_ancestor_12.fa,hg19_ancestral/homo_sapiens_ancestor_13.fa,hg19_ancestral/homo_sapiens_ancestor_14.fa,hg19_ancestral/homo_sapiens_ancestor_15.fa,hg19_ancestral/homo_sapiens_ancestor_16.fa,hg19_ancestral/homo_sapiens_ancestor_17.fa,hg19_ancestral/homo_sapiens_ancestor_18.fa,hg19_ancestral/homo_sapiens_ancestor_19.fa,hg19_ancestral/homo_sapiens_ancestor_20.fa,hg19_ancestral/homo_sapiens_ancestor_21.fa,hg19_ancestral/homo_sapiens_ancestor_22.fa,hg19_ancestral/homo_sapiens_ancestor_X.fa \
-refgenome=hg19_refgenome/chr1.fa,hg19_refgenome/chr2.fa,hg19_refgenome/chr3.fa,hg19_refgenome/chr4.fa,hg19_refgenome/chr5.fa,hg19_refgenome/chr6.fa,hg19_refgenome/chr7.fa,hg19_refgenome/chr8.fa,hg19_refgenome/chr9.fa,hg19_refgenome/chr10.fa,hg19_refgenome/chr11.fa,hg19_refgenome/chr12.fa,hg19_refgenome/chr13.fa,hg19_refgenome/chr14.fa,hg19_refgenome/chr15.fa,hg19_refgenome/chr16.fa,hg19_refgenome/chr17.fa,hg19_refgenome/chr18.fa,hg19_refgenome/chr19.fa,hg19_refgenome/chr20.fa,hg19_refgenome/chr21.fa,hg19_refgenome/chr22.fa,hg19_refgenome/chrX.fa \
-weights=strickmask.bed \
-out=outgroup.txt
Estimating mutation rate across genome
We can use the number of variants in the outgroup to estimate the substitution rate as a proxy for mutation rate.
(took 30 sec) > hmmix mutation_rate -outgroup=outgroup.txt -weights=strickmask.bed -window_size=1000000 -out mutationrate.bed
----------------------------------------
> Outgroupfile: outgroup.txt
> Outputfile is: mutationrate.bed
> Callability file is: strickmask.bed
> Window size: 1000000
----------------------------------------
Find a set of variants which are not derived in the outgroup
Keep variants that are not found to be derived in the outgroup for each individual in ingroup. You can also speficy a single individual or a comma separated list of individuals.
(took 20 min) > hmmix create_ingroup -ind=individuals.json -vcf=*.bcf -weights=strickmask.bed -out=obs -outgroup=outgroup.txt -ancestral=hg19_ancestral/homo_sapiens_ancestor_*.fa
----------------------------------------
> Ingroup individuals: 2
> Using vcf and ancestral files
vcffile: chr1.bcf ancestralfile: hg19_ancestral/homo_sapiens_ancestor_1.fa
vcffile: chr2.bcf ancestralfile: hg19_ancestral/homo_sapiens_ancestor_2.fa
vcffile: chr3.bcf ancestralfile: hg19_ancestral/homo_sapiens_ancestor_3.fa
vcffile: chr4.bcf ancestralfile: hg19_ancestral/homo_sapiens_ancestor_4.fa
vcffile: chr5.bcf ancestralfile: hg19_ancestral/homo_sapiens_ancestor_5.fa
vcffile: chr6.bcf ancestralfile: hg19_ancestral/homo_sapiens_ancestor_6.fa
vcffile: chr7.bcf ancestralfile: hg19_ancestral/homo_sapiens_ancestor_7.fa
vcffile: chr8.bcf ancestralfile: hg19_ancestral/homo_sapiens_ancestor_8.fa
vcffile: chr9.bcf ancestralfile: hg19_ancestral/homo_sapiens_ancestor_9.fa
vcffile: chr10.bcf ancestralfile: hg19_ancestral/homo_sapiens_ancestor_10.fa
vcffile: chr11.bcf ancestralfile: hg19_ancestral/homo_sapiens_ancestor_11.fa
vcffile: chr12.bcf ancestralfile: hg19_ancestral/homo_sapiens_ancestor_12.fa
vcffile: chr13.bcf ancestralfile: hg19_ancestral/homo_sapiens_ancestor_13.fa
vcffile: chr14.bcf ancestralfile: hg19_ancestral/homo_sapiens_ancestor_14.fa
vcffile: chr15.bcf ancestralfile: hg19_ancestral/homo_sapiens_ancestor_15.fa
vcffile: chr16.bcf ancestralfile: hg19_ancestral/homo_sapiens_ancestor_16.fa
vcffile: chr17.bcf ancestralfile: hg19_ancestral/homo_sapiens_ancestor_17.fa
vcffile: chr18.bcf ancestralfile: hg19_ancestral/homo_sapiens_ancestor_18.fa
vcffile: chr19.bcf ancestralfile: hg19_ancestral/homo_sapiens_ancestor_19.fa
vcffile: chr20.bcf ancestralfile: hg19_ancestral/homo_sapiens_ancestor_20.fa
vcffile: chr21.bcf ancestralfile: hg19_ancestral/homo_sapiens_ancestor_21.fa
vcffile: chr22.bcf ancestralfile: hg19_ancestral/homo_sapiens_ancestor_22.fa
vcffile: chrX.bcf ancestralfile: hg19_ancestral/homo_sapiens_ancestor_X.fa
> Using outgroup variants from: outgroup.txt
> Callability file: strickmask.bed
> Writing output to file with prefix: obs.<individual>.txt
----------------------------------------
Running command:
bcftools view -m2 -M2 -v snps -s HG00096 -T strickmask.bed chr1.bcf | vcftools --vcf - --exclude-positions outgroup.txt --recode --stdout
...
bcftools view -m2 -M2 -v snps -s HG00097 -T strickmask.bed chr22.bcf | vcftools --vcf - --exclude-positions outgroup.txt --recode --stdout
# Different way to define which individuals are in the ingroup
(took 20 min) > hmmix create_ingroup -ind=HG00096,HG00097 -vcf=*.bcf -weights=strickmask.bed -out=obs -outgroup=outgroup.txt -ancestral=hg19_ancestral/homo_sapiens_ancestor_*.fa
Training
Now for training the HMM parameters and decoding
(took 2 min) > hmmix train -obs=obs.HG00096.txt -weights=strickmask.bed -mutrates=mutationrate.bed -out=trained.HG00096.json
----------------------------------------
> state_names = ['Human', 'Archaic']
> starting_probabilities = [0.98, 0.02]
> transitions = [[1.0, 0.0], [0.02, 0.98]]
> emissions = [0.04, 0.4]
> chromosomes to use: All
> number of windows: 3032224 . Number of snps = 129803
> total callability: 0.72
> average mutation rate per bin: 1.0
> Output is trained.HG00096.json
> Window size is 1000 bp
> Haploid False
----------------------------------------
iteration loglikelihood start1 start2 emis1 emis2 trans1_1 trans2_2
0 -495665.7379 0.98 0.02 0.04 0.4 0.9999 0.98
1 -493092.9242 0.964 0.036 0.0459 0.3894 0.9995 0.9859
2 -492917.6235 0.959 0.041 0.0455 0.3847 0.9993 0.9834
...
20 -492775.4072 0.954 0.046 0.0442 0.3725 0.9989 0.9768
21 -492775.4058 0.954 0.046 0.0442 0.3725 0.9989 0.9768
22 -492775.4049 0.954 0.046 0.0442 0.3725 0.9989 0.9768
Decoding
(took 30 sec) > hmmix decode -obs=obs.HG00096.txt -weights=strickmask.bed -mutrates=mutationrate.bed -param=trained.HG00096.json
----------------------------------------
> state_names = ['Human', 'Archaic']
> starting_probabilities = [0.954, 0.046]
> transitions = [[0.999, 0.001], [0.023, 0.977]]
> emissions = [0.044, 0.372]
> chromosomes to use: All
> number of windows: 3032224 . Number of snps = 129803
> total callability: 0.72
> average mutation rate per bin: 1.0
> Output prefix is /dev/stdout
> Window size is 1000 bp
> Haploid False
----------------------------------------
chrom start end length state mean_prob snps
1 0 2988000 2988000 Human 0.98429 91
1 2988000 2997000 9000 Archaic 0.76217 6
1 2997000 3425000 428000 Human 0.98776 30
1 3425000 3452000 27000 Archaic 0.95816 22
1 3452000 4302000 850000 Human 0.97917 36
1 4302000 4361000 59000 Archaic 0.867 20
1 4361000 4500000 139000 Human 0.96854 4
1 4500000 4510000 10000 Archaic 0.8552 7
You can also save to an output file with the command:
hmmix decode -obs=obs.HG00096.txt -weights=strickmask.bed -mutrates=mutationrate.bed -param=trained.HG00096.json -out=HG00096.decoded
This will create a file named HG00096.decoded.diploid.txt because the default option is treating the data as diploid (more on haploid decoding in next chapter)
Training and decoding with phased data
It is also possible to tell the model that the data is phased with the -haploid parameter. For that we first need to train the parameters for haploid data and then decode. Training the model on phased data is done like this - and we also remember to change the name of the parameter file to include phased so future versions of ourselves don't forget. Another thing to note is that the number of snps is bigger than before 135483 vs 129803. This is because the program is counting snps on both haplotypes and homozygotes will be counted twice!
(took 4 min) > hmmix train -obs=obs.HG00096.txt -weights=strickmask.bed -mutrates=mutationrate.bed -out=trained.HG00096.phased.json -haploid
----------------------------------------
> state_names = ['Human', 'Archaic']
> starting_probabilities = [0.98, 0.02]
> transitions = [[1.0, 0.0], [0.02, 0.98]]
> emissions = [0.04, 0.4]
> chromosomes to use: All
> number of windows: 6064448 . Number of snps = 135483
> total callability: 0.72
> average mutation rate per bin: 1.0
> Output is trained.HG00096.phased.json
> Window size is 1000 bp
> Haploid True
----------------------------------------
iteration loglikelihood start1 start2 emis1 emis2 trans1_1 trans2_2
0 -605433.1928 0.98 0.02 0.04 0.4 0.9999 0.98
1 -589492.2 0.985 0.015 0.0248 0.3999 0.9998 0.9852
2 -588823.124 0.98 0.02 0.0238 0.3671 0.9996 0.9825
...
20 -588456.6027 0.973 0.027 0.0228 0.3267 0.9993 0.9755
21 -588456.6015 0.973 0.027 0.0228 0.3266 0.9993 0.9755
22 -588456.6009 0.973 0.027 0.0228 0.3266 0.9993 0.9755
Below I am only showing the first archaic segments on chromosome 1 for each haplotype (note you have to scroll down after chrom X before the new haplotype begins). The seem to fall more or less in the same places as when we used diploid data.
(took 30 sec) > hmmix decode -obs=obs.HG00096.txt -weights=strickmask.bed -mutrates=mutationrate.bed -param=trained.HG00096.phased.json -haploid
----------------------------------------
> state_names = ['Human', 'Archaic']
> starting_probabilities = [0.973, 0.027]
> transitions = [[0.999, 0.001], [0.024, 0.976]]
> emissions = [0.023, 0.327]
> chromosomes to use: All
> number of windows: 6064448 . Number of snps = 135483
> total callability: 0.72
> average mutation rate per bin: 1.0
> Output prefix is /dev/stdout
> Window size is 1000 bp
> Haploid True
----------------------------------------
hap1
chrom start end length state mean_prob snps
1 2156000 2185000 29000 Archaic 0.64711 6
1 3425000 3452000 27000 Archaic 0.96701 22
...
hap2
1 2780000 2803000 23000 Archaic 0.68285 7
1 4302000 4337000 35000 Archaic 0.94244 13
1 4500000 4511000 11000 Archaic 0.87938 7
1 4989000 5001000 12000 Archaic 0.61874 5
You can also save to an output file with the command:
hmmix decode -obs=obs.HG00096.txt -weights=strickmask.bed -mutrates=mutationrate.bed -param=trained.HG00096.phased.json -haploid -out=HG00096.decoded
This will create two files named HG00096.decoded.hap1.txt and HG00096.decoded.hap2.txt
Annotate with known admixing population
Even though this method does not use archaic reference genomes for finding segments you can still use them to annotate your segments. I have uploaded a VCF file containing 4 high coverage archaic genomes (3 Neanderthals and 1 Denisovan) here:
https://zenodo.org/records/7246376 (hg19 - the one I use in this example)
https://zenodo.org/records/13368126 (hg38)
If you have a vcf from the population that admixed in VCF/BCF format you can write this:
> hmmix decode -obs=obs.HG00096.txt -weights=strickmask.bed -mutrates=mutationrate.bed -param=trained.HG00096.json -admixpop=archaicvar/*bcf
----------------------------------------
> state_names = ['Human', 'Archaic']
> starting_probabilities = [0.954, 0.046]
> transitions = [[0.999, 0.001], [0.023, 0.977]]
> emissions = [0.044, 0.372]
> chromosomes to use: All
> number of windows: 3032224 . Number of snps = 129803
> total callability: 0.72
> average mutation rate per bin: 1.0
> Output prefix is /dev/stdout
> Window size is 1000 bp
> Haploid False
----------------------------------------
bcftools view -a -R obs.HG00096.txttemp archaicvar/highcov_ind_9.bcf
bcftools view -a -R obs.HG00096.txttemp archaicvar/highcov_ind_19.bcf
bcftools view -a -R obs.HG00096.txttemp archaicvar/highcov_ind_7.bcf
bcftools view -a -R obs.HG00096.txttemp archaicvar/highcov_ind_21.bcf
bcftools view -a -R obs.HG00096.txttemp archaicvar/highcov_ind_20.bcf
bcftools view -a -R obs.HG00096.txttemp archaicvar/highcov_ind_15.bcf
bcftools view -a -R obs.HG00096.txttemp archaicvar/highcov_ind_10.bcf
bcftools view -a -R obs.HG00096.txttemp archaicvar/highcov_ind_3.bcf
bcftools view -a -R obs.HG00096.txttemp archaicvar/highcov_ind_17.bcf
bcftools view -a -R obs.HG00096.txttemp archaicvar/highcov_ind_6.bcf
bcftools view -a -R obs.HG00096.txttemp archaicvar/highcov_ind_X.bcf
bcftools view -a -R obs.HG00096.txttemp archaicvar/highcov_ind_16.bcf
bcftools view -a -R obs.HG00096.txttemp archaicvar/highcov_ind_1.bcf
bcftools view -a -R obs.HG00096.txttemp archaicvar/highcov_ind_18.bcf
bcftools view -a -R obs.HG00096.txttemp archaicvar/highcov_ind_14.bcf
bcftools view -a -R obs.HG00096.txttemp archaicvar/highcov_ind_4.bcf
bcftools view -a -R obs.HG00096.txttemp archaicvar/highcov_ind_2.bcf
bcftools view -a -R obs.HG00096.txttemp archaicvar/highcov_ind_22.bcf
bcftools view -a -R obs.HG00096.txttemp archaicvar/highcov_ind_5.bcf
bcftools view -a -R obs.HG00096.txttemp archaicvar/highcov_ind_8.bcf
bcftools view -a -R obs.HG00096.txttemp archaicvar/highcov_ind_11.bcf
bcftools view -a -R obs.HG00096.txttemp archaicvar/highcov_ind_12.bcf
bcftools view -a -R obs.HG00096.txttemp archaicvar/highcov_ind_13.bcf
chrom start end length state mean_prob snps admixpopvariants AltaiNeandertal Vindija33.19 Denisova Chagyrskaya-Phalanx
1 2988000 2997000 9000 Archaic 0.76217 6 4 4 4 1 4
1 3425000 3452000 27000 Archaic 0.95816 22 17 17 15 3 17
1 4302000 4361000 59000 Archaic 0.867 20 12 11 12 11 11
1 4500000 4510000 10000 Archaic 0.8552 7 5 4 5 4 5
1 5306000 5319000 13000 Archaic 0.55584 4 1 1 1 0 1
1 5338000 5348000 10000 Archaic 0.65033 5 3 2 3 0 3
1 9321000 9355000 34000 Archaic 0.86414 9 0 0 0 0 0
1 12599000 12655000 56000 Archaic 0.91159 18 11 4 11 0 10
For the first segment there are 6 derived snps. Of these snps 4 are shared with Altai,Vindija, Denisova and Chagyrskaya. Only 1 is shared with Denisova so this segment likeli introgressed from Neanderthals
And that is it! Now you have run the model and gotten a set of parameters that you can interpret biologically (see my paper) and you have a list of segments that belong to the human and Archaic state.
If you have any questions about the use of the scripts, if you find errors or if you have feedback you can contact my here (make an issue) or write to: lauritsskov2@gmail.com
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
File details
Details for the file hmmix-0.8.1.tar.gz.
File metadata
- Download URL: hmmix-0.8.1.tar.gz
- Upload date:
- Size: 59.0 kB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/4.0.0 CPython/3.9.7
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | df3d5d8047a0ba02b9da5972356c26e08470dfffddbbbae10635b150e2c44781 |
|
| MD5 | effece386c5f4c4d649cd8b931f9e693 |
|
| BLAKE2b-256 | 287d858dd9c2aca10d62d448e156e9d7db4f9ea71310ebeac42de0cae0c9bb9e |
File details
Details for the file hmmix-0.8.1-py3-none-any.whl.
File metadata
- Download URL: hmmix-0.8.1-py3-none-any.whl
- Upload date:
- Size: 43.0 kB
- Tags: Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/4.0.0 CPython/3.9.7
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 829ebe5a6cdab497186783253ef004e42c7a66ba5c270f379215f9a7e861691f |
|
| MD5 | f7701759e74f9202baf582df68d9fd4e |
|
| BLAKE2b-256 | 31776222f01e2eda335ab7d1ba9d05c0ed9f64f801550edda342f3239d1614f5 |