Tools for calculating sequence properties of disordered proteins [from the Pappu Lab in at Washington University in St. Louis]
Project description
version 0.1.8 - March 2016
Introduction
localCIDER is a Python package developed by the lab of Rohit Pappu at Washington University in St. Louis for calculating and plotting parameters associated with intrinsically disordered proteins (IDPs) and disodered regions (IDRs). localCIDER is the Python backend for CIDER, ( Classification of Intrinsically Disordered Ensemble Regions) - a webserver currently in the final stages of development, also by the Pappu lab. Essentially, localCIDER lets you run CIDER’s calculations locally, allowing you to create custom analysis pipelines which do not rely on the webserver. It also allows you to take advantage of your own local infrastructure, rather than competing with everyone else for a common set of hardware provided by the Pappu lab.
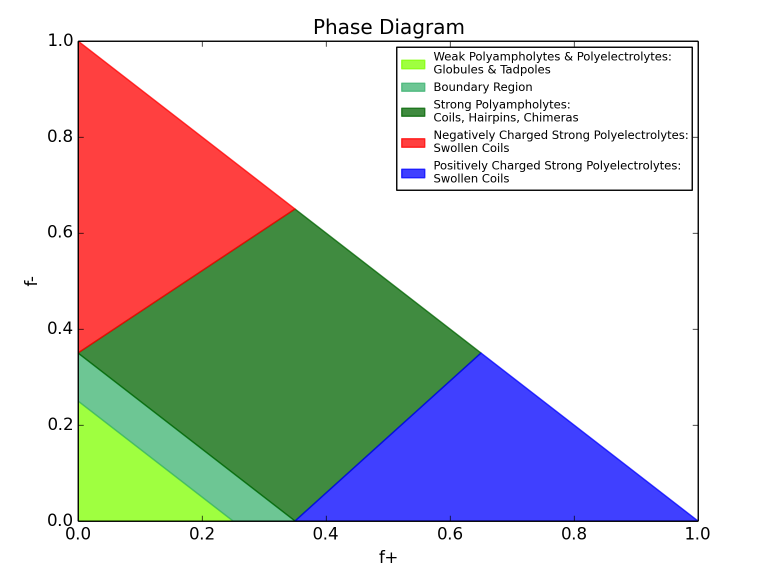
This project was motivated by the need to rapidly and easily calculate the κ (kappa) parameter, as defined in the 2013 Das & Pappu PNAS paper [1], as well as provide a tool to easily plot a sequence on the Das-Pappu diagram of states;
The original method for calculating κ involved an incredibly computationally expensive Monte Carlo step (used to determine the maximally segregated sequence) , whereas localCIDER makes use of a number of heuristics to reduce the Monte Carlo search space to an optimized and highly manageable size (most κ calculations are carried out in less than a second).
Beyond κ, localCIDER lets you calculate a range of additional parameters, as well as create the Das-Pappu diagram of states (shown below) and a number of other plots.
For more information please see the full documentation .
Installation
To install run
[sudo] pip install localcider
Note that localcider requires numpy and matplotlib to run.
Usage, bugs, and questions
Please see the see the full documentation for usage guidelines. Please address all questions and bug reports to Alex and he’ll do his best to get back to you!
About
localCIDER was written by Alex Holehouse and James Ahad in the Pappu Lab . A manuscript is currently under preparation for citation, but until that time please cite localCIDER as;
Holehouse, A. S., Ahad, J., Das, R. K. & Pappu, R. V. CIDER: Classification of Intrinsically Disordered Ensemble Regions. Biophys. J. 108, 228a (2015).
References
[1] Conformations of intrinsically disordered proteins are influenced by linear sequence distributions of oppositely charged residues R.K. Das & R.V. Pappu (2013) PNAS 110, 33, pp13392–13397.
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.











