mCodeGPT
Project description
mCodeGPT
Introduction
mCodeGPT is a Python package for the generation of cancer ontologies and Knowledge Bases using large language models (LLMs).
mCodeGPT makes use of so-called instruction prompts in LLMs such as GPT-4.
Two different strategies for knowledge extraction are currently implemented in mCodeGPT:
- SPIRES: Structured Prompt Interrogation and Recursive Extraction of Semantics
- A Zero-shot learning (ZSL) approach to extracting nested semantic structures from text
- This approach takes two inputs - 1) LinkML schema 2) free text, and outputs knowledge in a structure conformant with the supplied schema in JSON, YAML, RDF or OWL formats
- Uses GPT-3.5-turbo, GPT-4, or one of a variety of open LLMs on your local machine
- SPINDOCTOR: Structured Prompt Interpolation of Narrative Descriptions Or Controlled Terms for Ontological Reporting
- Summarize gene set descriptions (pseudo gene-set enrichment)
- Uses GPT-3.5-turbo or GPT-4
mCodeGPT is a redistribution with function modification of the python library ontogpt under BSD-3 license. Instructions below are mainly from the ontogpt but with modifications for cancer research.
Pre-requisites
-
Python 3.9+
-
OpenAI API key: necessary for using OpenAI's GPT models. This is a paid API and you will be charged based on usage. If you do not have an OpenAI account, you may sign up here. You will need to set your API key using the Ontology Access Kit:
poetry run runoak set-apikey -e openai <your openai api key>
You may also set additional API keys for optional resources:
- BioPortal account (for grounding). The BioPortal key is necessary for using ontologies from BioPortal. You may get a key by signing up for an account on their web site.
- NCBI E-utilities. The NCBI email address and API key are used for retrieving text and metadata from PubMed. You may still access these resources without identifying yourself, but you may encounter rate limiting and errors.
- HuggingFace Hub. This API key is necessary to retrieve models from the HuggingFace Hub service.
These optional keys may be set as follows:
poetry run runoak set-apikey -e bioportal <your bioportal api key>
poetry run runoak set-apikey -e ncbi-email <your email address>
poetry run runoak set-apikey -e ncbi-key <your NCBI api key>
poetry run runoak set-apikey -e hfhub-key <your HuggingFace Hub api key>
Setup
To simply start using the package in your workspace:
pip install mcodegpt
Note that some features require installing additional, optional dependencies.
These may be installed as:
poetry install --extras extra_name
# OR
pip install mcodegpt[extra_name]
where extra_name is one of the following:
docs- dependencies for building documentationweb- dependencies for the web applicationrecipes- dependencies for recipe scraping and parsingtextract- the textract pluginhuggingface- dependencies for accessing LLMs from HuggingFace Hub, remotely or locally
For feature development and contributing to the package:
git clone https://github.com/monarch-initiative/mcodegpt.git
cd ~/path/to/mcodegpt
poetry install
Getting Started
mCodeGPT is run from the command line. See the full list of commands with:
mcodegpt --help
For a simple example of text completion and testing to ensure mCodeGPT is set up correctly, create a text file containing the following, saving the file as example.txt:
Why did the squid cross the coral reef?
Then try the following command:
mcodegpt complete example.txt
You should get text output like the following:
Perhaps the squid crossed the coral reef for a variety of reasons:
1. Food: Squids are known to feed on small fish and other marine organisms, and there could have been a rich food source on the other side of the reef.
...
mCodeGPT is intended to be used for information extraction. The following examples show how to accomplish this.
Strategy 1: Knowledge extraction using SPIRES
Working Mechanism
- You provide an arbitrary data model, describing the structure you want to extract text into. This can be nested (but see limitations below). The predefined templates may be used.
- Provide your preferred annotations for grounding
NamedEntityfields - mCodeGPT will:
- Generate a prompt
- Feed the prompt to a language model
- Parse the results into a dictionary structure
- Ground the results using a preferred annotator (e.g., an ontology)
Input
Consider some text from one of the input files being used in the mCodeGPT test suite. You can find the text file here. You can download the raw file from the GitHub link to that input text file, or copy its contents over into another file, say, abstract.txt. An excerpt:
Patient John Smith (HSI-5421) underwent a tissue biopsy at the bladder collection site. The histopathology report confirmed the presence of cancer in the
... [cancer tumor size] ...
lives in zip code 90210. He is still alive, with no reported death date at this time. For any further information, please contact John Smith at jsmith@email.com or (555) 123-4567. ... ...
We can extract knowledge from the above text this into the FHIR mCode Cancer Ontology Standard by running the following command:
Command
mcodegpt extract -t cancer.CancerAnnotations -i ~/path/to/abstract.txt
The mCode standard is from HL7 FHIR.
Note: The value accepted by the -t / --template argument is the base name of one of the LinkML schema / data model which can be found in the templates folder.
Output
The output returned from the above command can be optionally redirected into an output file using the -o / --output.
The following is a small part of what the larger schema-compliant output looks like:
raw_completion_output: |-
human_specimen_identifier: HSI-5421
human_specimen_collection_site: bladder collection site
human_specimen_specimen_type: tissue biopsy
name: John Smith
contact_info: jsmith@email.com or (555) 123-4567
birth_date: April 15, 1975
gender: male
zip_code: 90210
us_core_race: Caucasian
us_core_birth_sex: male
us_core_ethnicity: non-Hispanic
death_date: N/A
disease_status_evidence_type: histopathology report
tumor_identifier: T-BC5421
tumor_body_location: bladder
tumor_size_longest_dimension: 4 cm
cancer_stage_stage_type: T2
cancer_asserted_date: January 10, 2022
cancer_body_site: bladder
tumor_marker_test_result_value: elevated levels of urinary bladder tumor antigen (UBTA)
people_human_specimen_identifier_interaction: John Smith
people_human_specimen_collection_site_interaction: bladder collection site
people_human_specimen_specimen_type_interaction: tissue biopsy
people_name_interaction: John Smith
people_contact_info_interaction: jsmith@email.com or (555) 123-4567
people_birth_date_interaction: April 15, 1975
people_gender_interaction: male
people_zip_code_interaction: 90210
people_us_core_race_interaction: Caucasian
people_us_core_birth_sex_interaction: male
people_us_core_ethnicity_interaction: non-Hispanic
people_death_date_interaction: N/A
people_disease_status_evidence_type_interaction: histopathology report
people_tumor_identifier_interaction: T-BC5421
people_tumor_body_location_interaction: bladder
people_tumor_size_longest_dimension_interaction: 4 cm
people_cancer_stage_stage_type_interaction: T2
people_cancer_asserted_date_interaction: January 10, 2022
people_cancer_body_site_interaction: bladder
people_tumor_marker_test_result_value_interaction: elevated levels of urinary bladder tumor antigen (UBTA)
Local Models
To use a local model, specify it with the -m or --model option.
Example:
mcodegpt extract -t drug -i ~/path/to/abstract.txt -m nous-hermes-13b
See the list of all available models with this command:
mcodegpt list-models
When specifying a local model for the first time, it will be downloaded to your local system.
Text length limit
LLMs have context sizes limiting the combined length of their inputs and outputs. The text-davinci-003 model, for example, whas a total 4,000 token limit (prompt + completion), while the gpt-3.5-turbo-16k model has a larger context of 16 thousand tokens.
Schema tips
It helps to have an understanding of the LinkML schema language, but it should be possible to define your own schemas using the examples in src/mcodegpt/templates as a guide.
mCodeGPT-specific extensions are specified as annotations.
You can specify a set of annotators for a field using the annotators annotation.
Ex.:
Gene:
is_a: NamedThing
id_prefixes:
- HGNC
annotations:
annotators: gilda:, bioportal:hgnc-nr, obo:pr
The annotators are applied in order.
Additionally, when performing grounding, the following measures can be taken to improve accuracy:
- Specify the valid set of ID prefixes using
id_prefixes - Some vocabularies have structural IDs that are amenable to regexes, you can specify these using
pattern - You can make use of
values_fromslot to specify a Dynamic Value Set- For example, you can constrain the set of valid locations for a gene product to be subclasses of
cellular_componentin GO orcellin CL
- For example, you can constrain the set of valid locations for a gene product to be subclasses of
Ex.:
classes:
...
GeneLocation:
is_a: NamedEntity
id_prefixes:
- GO
- CL
annotations:
annotators: "sqlite:obo:go, sqlite:obo:cl"
slot_usage:
id:
values_from:
- GOCellComponentType
- CellType
enums:
GOCellComponentType:
reachable_from:
source_ontology: obo:go
source_nodes:
- GO:0005575 ## cellular_component
CellType:
reachable_from:
source_ontology: obo:cl
source_nodes:
- CL:0000000 ## cell
OWL Exports
The extract command will let you export the results as OWL axioms, utilizing linkml-owl mappings in the schema.
Ex.:
mcodegpt extract -t recipe -i recipe-spaghetti.txt -o recipe-spaghetti.owl -O owl
src/mcodegpt/templates/recipe.yaml is an example schema that uses linkml-owl mappings.
See the Makefile for a full pipeline that involves using robot to extract a subset of FOODON and merge in the extracted results. This uses recipe-scrapers.
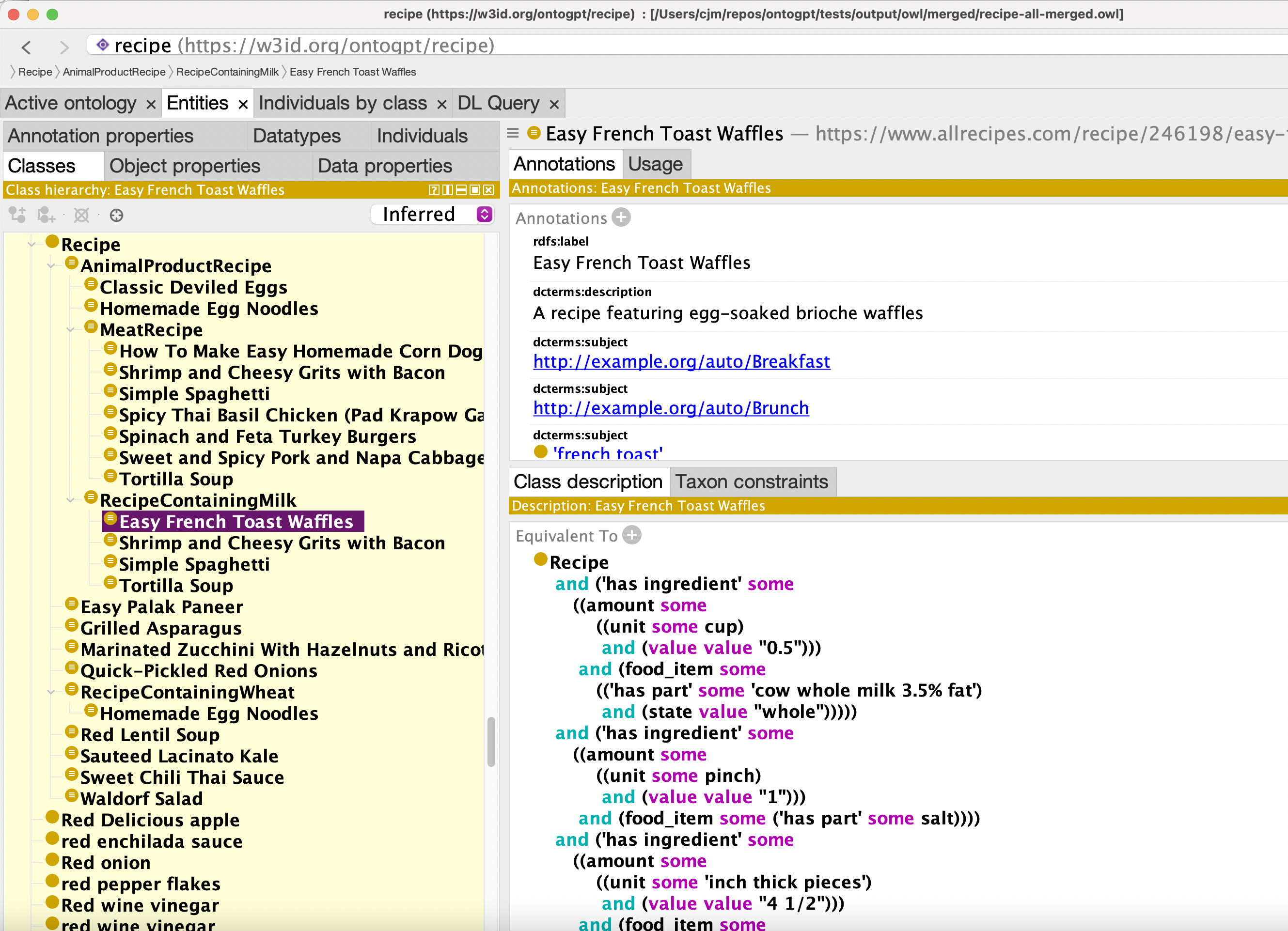
OWL output: recipe-all-merged.owl
Classification:
Web Application Setup
There is a bare bones web application for running mCodeGPT and viewing results.
Install the required dependencies by running the following command:
poetry install -E web
Then run this command to start the web application:
poetry run web-mcodegpt
Note: The agent running uvicorn must have the API key set, so for obvious reasons don't host this publicly without authentication, unless you want your credits drained.
SPINDOCTOR web app
To start:
poetry run streamlit run src/mcodegpt/streamlit/spindoctor.py
HuggingFace Hub
Note: support for HuggingFace-provided models is currently a work in progress.
A select number of LLMs may be accessed through HuggingFace Hub. See the full list using mcodegpt list-models
Specify a model name with the -m option.
Example:
mcodegpt extract -t mendelian_disease.MendelianDisease -i tests/input/cases/mendelian-disease-sly.txt -m FLAN_T5_BASE
Citation
SPIRES is described further in: Caufield JH, Hegde H, Emonet V, Harris NL, Joachimiak MP, Matentzoglu N, et al. Structured prompt interrogation and recursive extraction of semantics (SPIRES): A method for populating knowledge bases using zero-shot learning. arXiv publication: http://arxiv.org/abs/2304.02711
SPINDOCTOR is described further in: Joachimiak MP, Caufield JH, Harris NL, Kim H, Mungall CJ. Gene Set Summarization using Large Language Models. arXiv publication: http://arxiv.org/abs/2305.13338
Contributing
Contributions on recipes to test welcome from anyone.
Project details
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
Filter files by name, interpreter, ABI, and platform.
If you're not sure about the file name format, learn more about wheel file names.
Copy a direct link to the current filters
File details
Details for the file mcodegpt-0.2.25.tar.gz.
File metadata
- Download URL: mcodegpt-0.2.25.tar.gz
- Upload date:
- Size: 1.0 MB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/4.0.2 CPython/3.9.1
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
4880d3ee1f72f5c4c1437f89f8ae8b65231190574d9a6537db9b02148da4c279
|
|
| MD5 |
ca3b872c5ed2ec9a63d3da67e4ff7967
|
|
| BLAKE2b-256 |
74648565f59dce8d24768ff53fac0d2fabe86687f2455fff61d176efb173d2de
|
File details
Details for the file mcodegpt-0.2.25-py3-none-any.whl.
File metadata
- Download URL: mcodegpt-0.2.25-py3-none-any.whl
- Upload date:
- Size: 1.1 MB
- Tags: Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/4.0.2 CPython/3.9.1
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
a85492ec10cff2d6569a5ca750a6b71cbbbe2479378b90ebe706e11cfcddb878
|
|
| MD5 |
8fe857ad684ef8ac5d03ed4d78a3ff6b
|
|
| BLAKE2b-256 |
21a9d0d3c3417005e49e33629fd72cde90349361bc06817321535168f171235e
|












