membrane segmentation in 3D for cryo-ET
Project description
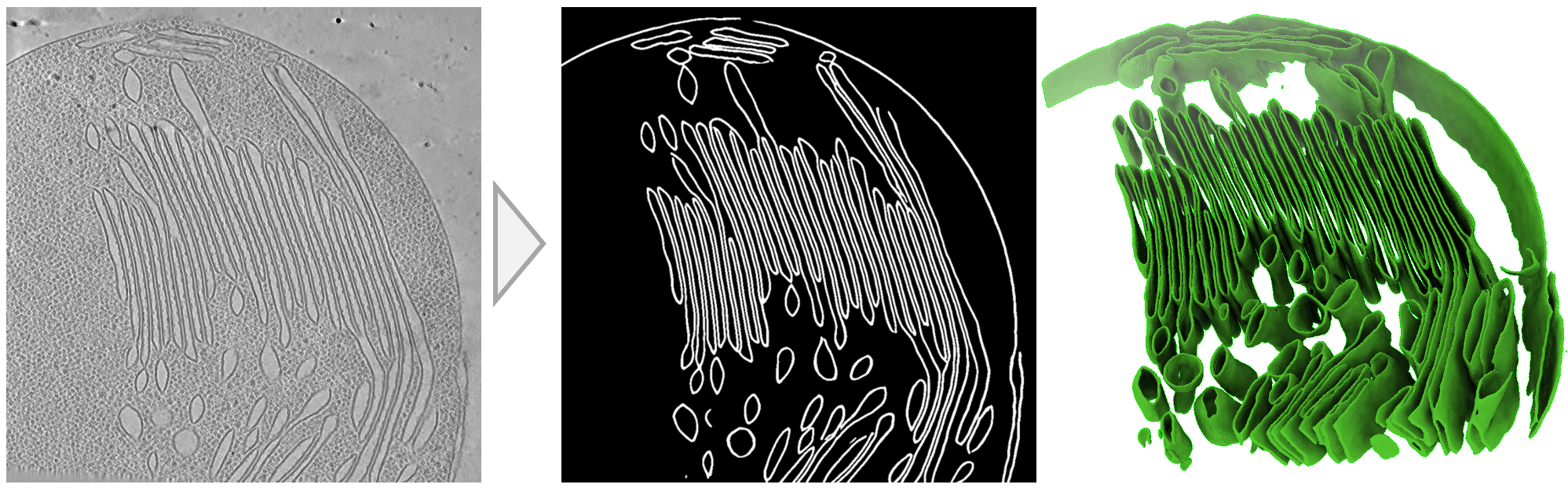
MemBrain-Seg
Membrain-Seg1 is a Python project developed by teamtomo for membrane segmentation in 3D for cryo-electron tomography (cryo-ET). This tool aims to provide researchers with an efficient and reliable method for segmenting membranes in 3D microscopic images. Membrain-Seg is currently under early development, so we may make breaking changes between releases.
Example notebook
For a quick start, you can walk through our example notebook. You can easily run it on Google Colab by clicking on the badge below:
Publication:
Membrain-seg's current functionalities are described on more detail in our preprint.
Membrain-Seg is currently under early development, so we may make breaking changes between releases.
Version Updates
For a detailed history of changes and updates, please refer to our CHANGELOG.md.
Overview
MemBrain-seg is a practical tool for membrane segmentation in cryo-electron tomograms. It's built on the U-Net architecture and makes use of a pre-trained model for efficient performance. The U-Net architecture and training parameters are largely inspired by nnUNet2.
Our current best model is available for download here. Please let us know how it works for you. If the given model does not work properly, you may want to try one of our experimental or previous versions:
Experimental models:
- v10_beta_FAaug -- model traing with Fourier amplitude augmentation for better generalization
- v10_beta_MWaug -- model traing with missing wedge augmentation for better missing wedge restoration
Other (older) model versions:
- v10_alpha -- standard model until 24th April 2025
- v9 -- best model until 10th Aug 2023
- v9b -- model for non-denoised data until 10th Aug 2023
If you wish, you can also train a new model using your own data, or combine it with our (soon to come!) publicly-available dataset.
To enhance segmentation, MemBrain-seg includes preprocessing functions. These help to adjust your tomograms so they're similar to the data our network was trained on, making the process smoother and more efficient.
Explore MemBrain-seg, use it for your needs, and let us know how it works for you!
Preliminary documentation is available, but far from perfect. Please let us know if you encounter any issues, and we are more than happy to help (and get feedback what does not work yet).
[1] Lamm, L., Zufferey, S., Righetto, R.D., Wietrzynski, W., Yamauchi, K.A., Burt, A., Liu, Y., Zhang, H., Martinez-Sanchez, A., Ziegler, S., Isensee, F., Schnabel, J.A., Engel, B.D., and Peng, T, 2024. MemBrain v2: an end-to-end tool for the analysis of membranes in cryo-electron tomography. bioRxiv, https://doi.org/10.1101/2024.01.05.574336
[2] Isensee, F., Jaeger, P.F., Kohl, S.A.A., Petersen, J., Maier-Hein, K.H., 2021. nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nature Methods 18, 203-211. https://doi.org/10.1038/s41592-020-01008-z
Installation
For detailed installation instructions, please look here.
Features
Segmentation
Segmenting the membranes in your tomograms is the main feature of this repository. Please find more detailed instructions here.
Preprocessing
Currently, we provide the following two preprocessing options:
- Pixel size matching: Rescale your tomogram to match the training pixel sizes
- Fourier amplitude matching: Scale Fourier components to match the "style" of different tomograms
- Deconvolution: denoises the tomogram by applying the deconvolution filter from Warp
For more information, see the Preprocessing subsection.
Model training
It is also possible to use this package to train your own model. Instructions can be found here.
Patch annotations
In case you would like to train a model that works better for your tomograms, it may be beneficial to add some more patches from your tomograms to the training dataset. Recommendations on how to to this can be found here.
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
Filter files by name, interpreter, ABI, and platform.
If you're not sure about the file name format, learn more about wheel file names.
Copy a direct link to the current filters
File details
Details for the file membrain_seg-0.0.10.tar.gz.
File metadata
- Download URL: membrain_seg-0.0.10.tar.gz
- Upload date:
- Size: 97.6 kB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/6.1.0 CPython/3.13.7
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
f770c0976b1a3370291b4a6f4d8cb9f83bbd65cab2bc676ce873ec559b2281a3
|
|
| MD5 |
117f8d6fa0001ebca2b886382b0c2b2c
|
|
| BLAKE2b-256 |
fd71015dbde6e1f275cc2fff57281285cb238c4125622a4e3442d0cbe8169753
|
File details
Details for the file membrain_seg-0.0.10-py3-none-any.whl.
File metadata
- Download URL: membrain_seg-0.0.10-py3-none-any.whl
- Upload date:
- Size: 110.8 kB
- Tags: Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/6.1.0 CPython/3.13.7
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
5b353a3e06507e477bb0d423c1181dcfe058f9bcc226cc4a0370cd5d79ce00e2
|
|
| MD5 |
3c5dc8833a25a2a52916c7fa7eb936cc
|
|
| BLAKE2b-256 |
9bfc6c3040506056005aa5dee8b7e46654d062016c6dbfc9ab454eb3ccba142a
|

















