Easy benchmarking of machine learning models with sklearn interface with statistical tests built-in.
Project description
Easy benchmarking of machine learning models with sklearn interface with statistical tests built-in.
Usage for classification problems
First, we consider the plot_classifier_comparison.py demo file. This extends the standard sklearn classifier comparison but also demos the ease of mlpaper to create a performance report.
In this demo, we use the example of the three toy data sets and ten classifiers from the sklearn example:
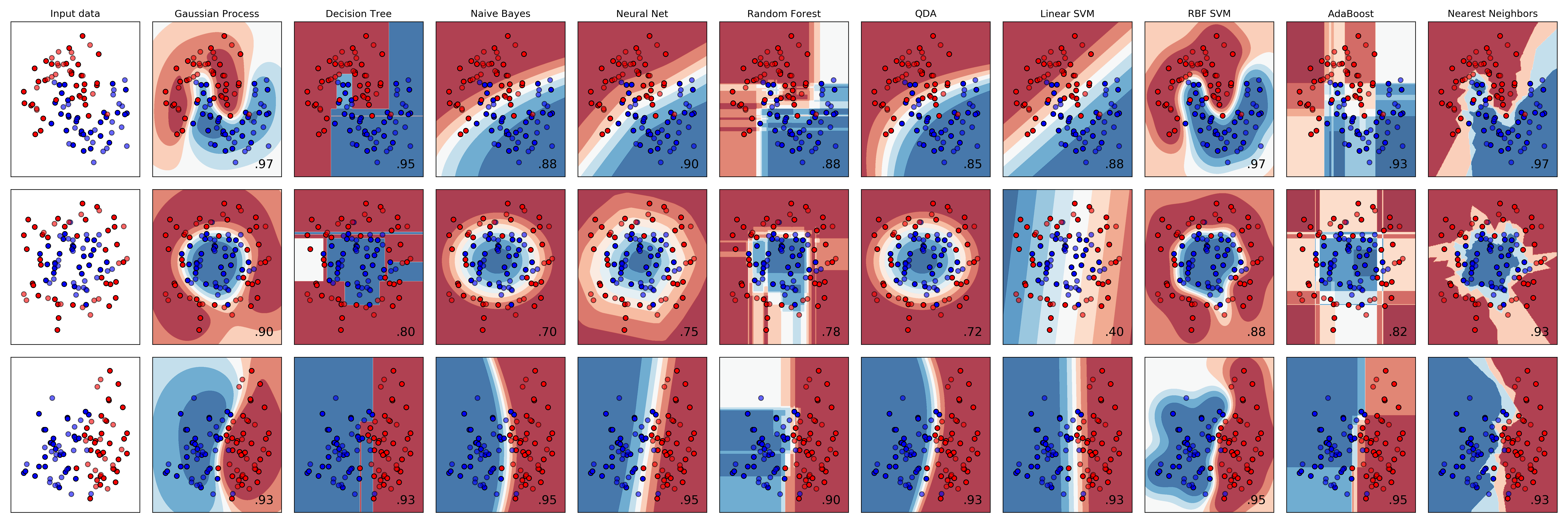
The mlpaper package can benchmark all of the of these methods and created a properly formatted LaTeX table (with error bars) in a few commands. This generates a results table for copy-and-paste into a ML paper .tex file in a few commands.
Pandas tables with the performance results of all the methods can be built by:
import mlpaper.classification as btc
from mlpaper.classification import STD_BINARY_CURVES, STD_CLASS_LOSS
performance_df, performance_curves_dict = btc.just_benchmark(
X_train,
y_train,
X_test,
y_test,
2,
classifiers,
STD_CLASS_LOSS,
STD_BINARY_CURVES,
ref_method,
)This benchmarks all the models in classifiers on the data (X_train, y_train, X_test, y_test) for 2-class classification. It uses the loss function described in the dictionaries STD_CLASS_LOSS, and the curves (e.g., ROC, PR) in STD_BINARY_CURVES. The ref_method defines the model that is the reference to compare against for assessing statistically significant performance gains.
The sciprint module formats these tables for scientific presentation. The performance dictionaries can be converted to cleanly formatted tables: correct significant figures, shifting of exponent for compactness, thresholding huge/small (crap limit) results, and correct alignment of decimal points, units in headers, etc. Here we use:
import mlpaper.sciprint as sp
print(
sp.just_format_it(
performance_df,
shift_mod=3,
unit_dict={"NLL": "nats"},
crap_limit_min={"AUPRG": -1},
EB_limit={"AUPRG": -1},
non_finite_fmt={sp.NAN_STR: "N/A"},
use_tex=False,
)
)to export the results in plain text, or for LaTeX we use:
import mlpaper.sciprint as sp
print(
sp.just_format_it(
performance_df,
shift_mod=3,
unit_dict={"NLL": "nats"},
crap_limit_min={"AUPRG": -1},
EB_limit={"AUPRG": -1},
non_finite_fmt={sp.NAN_STR: "{--}"},
use_tex=True,
)
)Output
Dataset 0 (Moons)
AP p AUC p AUPRG p Brier p NLL (nats) p sphere p zero one p AdaBoost 0.93(16) <0.0001 0.950(96) <0.0001 0.90464 <0.0001 0.42(14) <0.0001 0.368(80) <0.0001 0.36(15) <0.0001 0.075(86) <0.0001 Decision Tree 0.95(13) <0.0001 0.966(70) <0.0001 0.93860 <0.0001 0.18(25) <0.0001 0.40(71) 0.4072 0.16(22) <0.0001 0.050(71) <0.0001 Gaussian Process 0.90(22) <0.0001 0.95(12) <0.0001 0.92081 <0.0001 0.27(17) <0.0001 0.27(11) <0.0001 0.22(16) <0.0001 0.025(51) <0.0001 Linear SVM 0.952(99) <0.0001 0.950(77) <0.0001 0.88705 <0.0001 0.34(24) <0.0001 0.29(16) <0.0001 0.31(24) <0.0001 0.15(12) 0.0006 Naive Bayes 0.957(97) <0.0001 0.957(68) <0.0001 0.89782 <0.0001 0.34(25) <0.0001 0.28(18) <0.0001 0.31(24) <0.0001 0.13(11) 0.0002 Nearest Neighbors 0.94(14) <0.0001 0.969(69) <0.0001 0.93498 <0.0001 0.18(21) <0.0001 0.42(70) 0.4241 0.15(18) <0.0001 0.025(51) <0.0001 Neural Net 0.957(91) <0.0001 0.957(69) <0.0001 0.89782 <0.0001 0.33(23) <0.0001 0.28(15) <0.0001 0.30(22) <0.0001 0.100(98) <0.0001 QDA 0.951(91) <0.0001 0.950(80) <0.0001 0.88517 <0.0001 0.34(27) <0.0001 0.29(21) 0.0003 0.31(25) <0.0001 0.15(12) 0.0006 RBF SVM 0.93(18) <0.0001 0.957(94) <0.0001 0.92081 <0.0001 0.14(20) <0.0001 0.18(18) <0.0001 0.12(17) <0.0001 0.025(51) <0.0001 Random Forest 0.965(82) <0.0001 0.949(84) <0.0001 0.92147 <0.0001 0.31(26) <0.0001 0.52(70) 0.6099 0.28(24) <0.0001 0.100(98) <0.0001 iid 0.53(16) N/A 0.5(0) N/A 0(0) N/A 1.004(22) N/A 0.695(11) N/A 1.005(27) N/A 0.53(17) N/A
Dataset 0 (Moons) in LaTeX
\begin{tabular}{|l|Sr|Sr|Sr|Sr|Sr|Sr|Sr|}
\toprule
{} & {AP} & {p} & {AUC} & {p} & {AUPRG} & {p} & {Brier} & {p} & {NLL (nats)} & {p} & {sphere} & {p} & {zero one} & {p} \\
\midrule
AdaBoost & 0.93(16) & <0.0001 & 0.950(96) & <0.0001 & 0.90464 & <0.0001 & 0.42(14) & <0.0001 & 0.368(80) & <0.0001 & 0.36(15) & <0.0001 & 0.075(86) & <0.0001 \\
Decision Tree & 0.95(13) & <0.0001 & 0.966(70) & <0.0001 & 0.93860 & <0.0001 & 0.18(25) & <0.0001 & 0.40(71) & 0.4072 & 0.16(22) & <0.0001 & 0.050(71) & <0.0001 \\
Gaussian Process & 0.90(22) & <0.0001 & 0.95(12) & <0.0001 & 0.92081 & <0.0001 & 0.27(17) & <0.0001 & 0.27(11) & <0.0001 & 0.22(16) & <0.0001 & 0.025(51) & <0.0001 \\
Linear SVM & 0.952(99) & <0.0001 & 0.950(77) & <0.0001 & 0.88705 & <0.0001 & 0.34(24) & <0.0001 & 0.29(16) & <0.0001 & 0.31(24) & <0.0001 & 0.15(12) & 0.0006 \\
Naive Bayes & 0.957(97) & <0.0001 & 0.957(68) & <0.0001 & 0.89782 & <0.0001 & 0.34(25) & <0.0001 & 0.28(18) & <0.0001 & 0.31(24) & <0.0001 & 0.13(11) & 0.0002 \\
Nearest Neighbors & 0.94(14) & <0.0001 & 0.969(69) & <0.0001 & 0.93498 & <0.0001 & 0.18(21) & <0.0001 & 0.42(70) & 0.4241 & 0.15(18) & <0.0001 & 0.025(51) & <0.0001 \\
Neural Net & 0.957(91) & <0.0001 & 0.957(69) & <0.0001 & 0.89782 & <0.0001 & 0.33(23) & <0.0001 & 0.28(15) & <0.0001 & 0.30(22) & <0.0001 & 0.100(98) & <0.0001 \\
QDA & 0.951(91) & <0.0001 & 0.950(80) & <0.0001 & 0.88517 & <0.0001 & 0.34(27) & <0.0001 & 0.29(21) & 0.0003 & 0.31(25) & <0.0001 & 0.15(12) & 0.0006 \\
RBF SVM & 0.93(18) & <0.0001 & 0.957(94) & <0.0001 & 0.92081 & <0.0001 & 0.14(20) & <0.0001 & 0.18(18) & <0.0001 & 0.12(17) & <0.0001 & 0.025(51) & <0.0001 \\
Random Forest & 0.965(82) & <0.0001 & 0.949(84) & <0.0001 & 0.92147 & <0.0001 & 0.31(26) & <0.0001 & 0.52(70) & 0.6099 & 0.28(24) & <0.0001 & 0.100(98) & <0.0001 \\
iid & 0.53(16) & {--} & 0.5(0) & {--} & 0(0) & {--} & 1.004(22) & {--} & 0.695(11) & {--} & 1.005(27) & {--} & 0.53(17) & {--} \\
\bottomrule
\end{tabular}
Dataset 1 (Circles)
AP p AUC p AUPRG p Brier p NLL (nats) p sphere p zero one p AdaBoost 0.938(82) <0.0001 0.89(12) <0.0001 0.76091 <0.0001 0.773(96) <0.0001 0.576(50) <0.0001 0.73(12) <0.0001 0.17(13) <0.0001 Decision Tree 0.86(16) <0.0001 0.80(13) <0.0001 0.76316 <0.0001 0.80(52) 0.3009 2.8(18) 0.0270 0.68(45) 0.0792 0.20(13) 0.0003 Gaussian Process 0.977(47) <0.0001 0.964(60) <0.0001 0.93049 <0.0001 0.39(23) <0.0001 0.33(14) <0.0001 0.36(23) <0.0001 0.100(98) <0.0001 Linear SVM 0.53(18) 0.1621 0.51(21) 0.8580 0.19756 0.3660 1.066(80) 0.1521 0.726(41) 0.1514 1.079(96) 0.1531 0.60(16) 1.0000 Naive Bayes 0.9983(82) <0.0001 0.997(13) <0.0001 0.996(21) <0.0001 0.64(20) <0.0001 0.48(12) <0.0001 0.63(21) <0.0001 0.30(15) 0.0003 Nearest Neighbors 0.996(15) <0.0001 0.966(49) <0.0001 0.991(47) <0.0001 0.30(16) <0.0001 0.23(11) <0.0001 0.28(16) <0.0001 0.075(86) <0.0001 Neural Net 0.993(23) <0.0001 0.990(32) <0.0001 0.982(79) <0.0001 0.69(14) <0.0001 0.525(74) <0.0001 0.65(16) <0.0001 0.25(15) <0.0001 QDA 0.9983(83) <0.0001 0.997(11) <0.0001 0.996(32) <0.0001 0.63(19) <0.0001 0.47(11) <0.0001 0.61(20) <0.0001 0.28(15) <0.0001 RBF SVM 0.979(44) <0.0001 0.966(63) <0.0001 0.93680 <0.0001 0.34(22) <0.0001 0.29(14) <0.0001 0.31(22) <0.0001 0.100(98) <0.0001 Random Forest 0.90(13) <0.0001 0.85(16) <0.0001 0.64512 0.0021 0.65(30) 0.0070 0.48(19) 0.0094 0.62(31) 0.0047 0.23(14) 0.0006 iid 0.60(16) N/A 0.5(0) N/A 0(0) N/A 1.071(85) N/A 0.729(43) N/A 1.08(11) N/A 0.60(16) N/A
Dataset 1 (Circles) in LaTeX
\begin{tabular}{|l|Sr|Sr|Sr|Sr|Sr|Sr|Sr|}
\toprule
{} & {AP} & {p} & {AUC} & {p} & {AUPRG} & {p} & {Brier} & {p} & {NLL (nats)} & {p} & {sphere} & {p} & {zero one} & {p} \\
\midrule
AdaBoost & 0.938(82) & <0.0001 & 0.89(12) & <0.0001 & 0.76091 & <0.0001 & 0.773(96) & <0.0001 & 0.576(50) & <0.0001 & 0.73(12) & <0.0001 & 0.17(13) & <0.0001 \\
Decision Tree & 0.86(16) & <0.0001 & 0.80(13) & <0.0001 & 0.76316 & <0.0001 & 0.80(52) & 0.3009 & 2.8(18) & 0.0270 & 0.68(45) & 0.0792 & 0.20(13) & 0.0003 \\
Gaussian Process & 0.977(47) & <0.0001 & 0.964(60) & <0.0001 & 0.93049 & <0.0001 & 0.39(23) & <0.0001 & 0.33(14) & <0.0001 & 0.36(23) & <0.0001 & 0.100(98) & <0.0001 \\
Linear SVM & 0.53(18) & 0.1621 & 0.51(21) & 0.8580 & 0.19756 & 0.3660 & 1.066(80) & 0.1521 & 0.726(41) & 0.1514 & 1.079(96) & 0.1531 & 0.60(16) & 1.0000 \\
Naive Bayes & 0.9983(82) & <0.0001 & 0.997(13) & <0.0001 & 0.996(21) & <0.0001 & 0.64(20) & <0.0001 & 0.48(12) & <0.0001 & 0.63(21) & <0.0001 & 0.30(15) & 0.0003 \\
Nearest Neighbors & 0.996(15) & <0.0001 & 0.966(49) & <0.0001 & 0.991(47) & <0.0001 & 0.30(16) & <0.0001 & 0.23(11) & <0.0001 & 0.28(16) & <0.0001 & 0.075(86) & <0.0001 \\
Neural Net & 0.993(23) & <0.0001 & 0.990(32) & <0.0001 & 0.982(79) & <0.0001 & 0.69(14) & <0.0001 & 0.525(74) & <0.0001 & 0.65(16) & <0.0001 & 0.25(15) & <0.0001 \\
QDA & 0.9983(83) & <0.0001 & 0.997(11) & <0.0001 & 0.996(32) & <0.0001 & 0.63(19) & <0.0001 & 0.47(11) & <0.0001 & 0.61(20) & <0.0001 & 0.28(15) & <0.0001 \\
RBF SVM & 0.979(44) & <0.0001 & 0.966(63) & <0.0001 & 0.93680 & <0.0001 & 0.34(22) & <0.0001 & 0.29(14) & <0.0001 & 0.31(22) & <0.0001 & 0.100(98) & <0.0001 \\
Random Forest & 0.90(13) & <0.0001 & 0.85(16) & <0.0001 & 0.64512 & 0.0021 & 0.65(30) & 0.0070 & 0.48(19) & 0.0094 & 0.62(31) & 0.0047 & 0.23(14) & 0.0006 \\
iid & 0.60(16) & {--} & 0.5(0) & {--} & 0(0) & {--} & 1.071(85) & {--} & 0.729(43) & {--} & 1.08(11) & {--} & 0.60(16) & {--} \\
\bottomrule
\end{tabular}
Dataset 2 (Linear)
AP p AUC p AUPRG p Brier p NLL (nats) p sphere p zero one p AdaBoost 0.984(43) <0.0001 0.962(87) <0.0001 0.96274 <0.0001 0.21(23) <0.0001 0.27(29) 0.0034 0.18(20) <0.0001 0.050(71) <0.0001 Decision Tree 0.91(14) <0.0001 0.922(98) <0.0001 0.88360 <0.0001 0.30(35) 0.0002 1.0(12) 0.5706 0.26(30) <0.0001 0.075(86) <0.0001 Gaussian Process 0.984(38) <0.0001 0.977(52) <0.0001 0.96794 <0.0001 0.25(24) <0.0001 0.23(17) <0.0001 0.23(23) <0.0001 0.075(86) <0.0001 Linear SVM 0.994(26) <0.0001 0.992(23) <0.0001 0.989(47) <0.0001 0.17(14) <0.0001 0.163(86) <0.0001 0.16(15) <0.0001 0.050(71) <0.0001 Naive Bayes 0.992(25) <0.0001 0.990(32) <0.0001 0.986(50) <0.0001 0.18(20) <0.0001 0.15(15) <0.0001 0.17(19) <0.0001 0.050(71) <0.0001 Nearest Neighbors 0.992(25) <0.0001 0.946(78) <0.0001 0.985(67) <0.0001 0.29(30) <0.0001 0.76(98) 0.9063 0.25(26) <0.0001 0.075(86) <0.0001 Neural Net 0.987(35) <0.0001 0.982(40) <0.0001 0.975(83) <0.0001 0.24(19) <0.0001 0.22(12) <0.0001 0.21(19) <0.0001 0.050(71) <0.0001 QDA 0.984(42) <0.0001 0.975(57) <0.0001 0.96560 <0.0001 0.21(24) <0.0001 0.23(28) 0.0014 0.19(22) <0.0001 0.075(86) <0.0001 RBF SVM 0.980(45) <0.0001 0.970(62) <0.0001 0.95778 <0.0001 0.21(25) <0.0001 0.20(21) <0.0001 0.18(23) <0.0001 0.050(71) <0.0001 Random Forest 0.990(25) <0.0001 0.968(58) <0.0001 0.981(73) <0.0001 0.25(25) <0.0001 0.47(70) 0.5055 0.23(23) <0.0001 0.075(86) <0.0001 iid 0.55(16) N/A 0.5(0) N/A 0(0) N/A 1.018(43) N/A 0.702(22) N/A 1.021(52) N/A 0.55(17) N/A
Dataset 2 (Linear) in LaTeX
\begin{tabular}{|l|Sr|Sr|Sr|Sr|Sr|Sr|Sr|}
\toprule
{} & {AP} & {p} & {AUC} & {p} & {AUPRG} & {p} & {Brier} & {p} & {NLL (nats)} & {p} & {sphere} & {p} & {zero one} & {p} \\
\midrule
AdaBoost & 0.984(43) & <0.0001 & 0.962(87) & <0.0001 & 0.96274 & <0.0001 & 0.21(23) & <0.0001 & 0.27(29) & 0.0034 & 0.18(20) & <0.0001 & 0.050(71) & <0.0001 \\
Decision Tree & 0.91(14) & <0.0001 & 0.922(98) & <0.0001 & 0.88360 & <0.0001 & 0.30(35) & 0.0002 & 1.0(12) & 0.5706 & 0.26(30) & <0.0001 & 0.075(86) & <0.0001 \\
Gaussian Process & 0.984(38) & <0.0001 & 0.977(52) & <0.0001 & 0.96794 & <0.0001 & 0.25(24) & <0.0001 & 0.23(17) & <0.0001 & 0.23(23) & <0.0001 & 0.075(86) & <0.0001 \\
Linear SVM & 0.994(26) & <0.0001 & 0.992(23) & <0.0001 & 0.989(47) & <0.0001 & 0.17(14) & <0.0001 & 0.163(86) & <0.0001 & 0.16(15) & <0.0001 & 0.050(71) & <0.0001 \\
Naive Bayes & 0.992(25) & <0.0001 & 0.990(32) & <0.0001 & 0.986(50) & <0.0001 & 0.18(20) & <0.0001 & 0.15(15) & <0.0001 & 0.17(19) & <0.0001 & 0.050(71) & <0.0001 \\
Nearest Neighbors & 0.992(25) & <0.0001 & 0.946(78) & <0.0001 & 0.985(67) & <0.0001 & 0.29(30) & <0.0001 & 0.76(98) & 0.9063 & 0.25(26) & <0.0001 & 0.075(86) & <0.0001 \\
Neural Net & 0.987(35) & <0.0001 & 0.982(40) & <0.0001 & 0.975(83) & <0.0001 & 0.24(19) & <0.0001 & 0.22(12) & <0.0001 & 0.21(19) & <0.0001 & 0.050(71) & <0.0001 \\
QDA & 0.984(42) & <0.0001 & 0.975(57) & <0.0001 & 0.96560 & <0.0001 & 0.21(24) & <0.0001 & 0.23(28) & 0.0014 & 0.19(22) & <0.0001 & 0.075(86) & <0.0001 \\
RBF SVM & 0.980(45) & <0.0001 & 0.970(62) & <0.0001 & 0.95778 & <0.0001 & 0.21(25) & <0.0001 & 0.20(21) & <0.0001 & 0.18(23) & <0.0001 & 0.050(71) & <0.0001 \\
Random Forest & 0.990(25) & <0.0001 & 0.968(58) & <0.0001 & 0.981(73) & <0.0001 & 0.25(25) & <0.0001 & 0.47(70) & 0.5055 & 0.23(23) & <0.0001 & 0.075(86) & <0.0001 \\
iid & 0.55(16) & {--} & 0.5(0) & {--} & 0(0) & {--} & 1.018(43) & {--} & 0.702(22) & {--} & 1.021(52) & {--} & 0.55(17) & {--} \\
\bottomrule
\end{tabular}
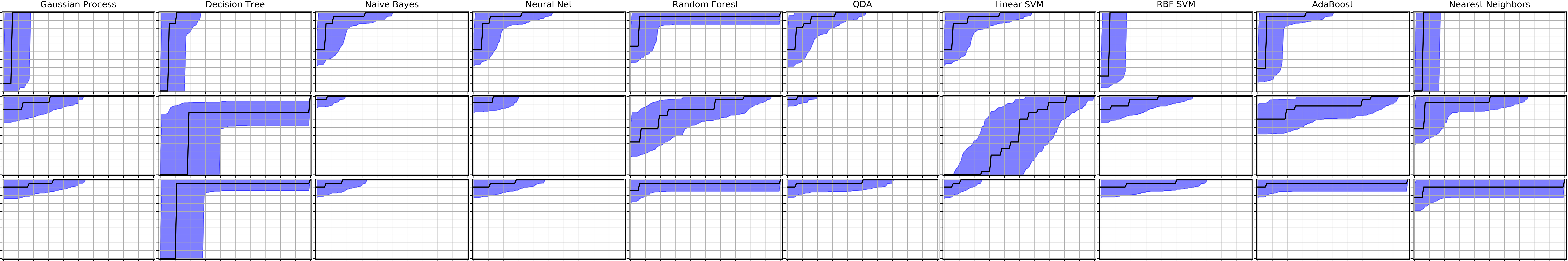
ROC curves
The just_benchmark routines also produces ROC curves with error bars from bootstrap analysis, which have been vectorized for speed:
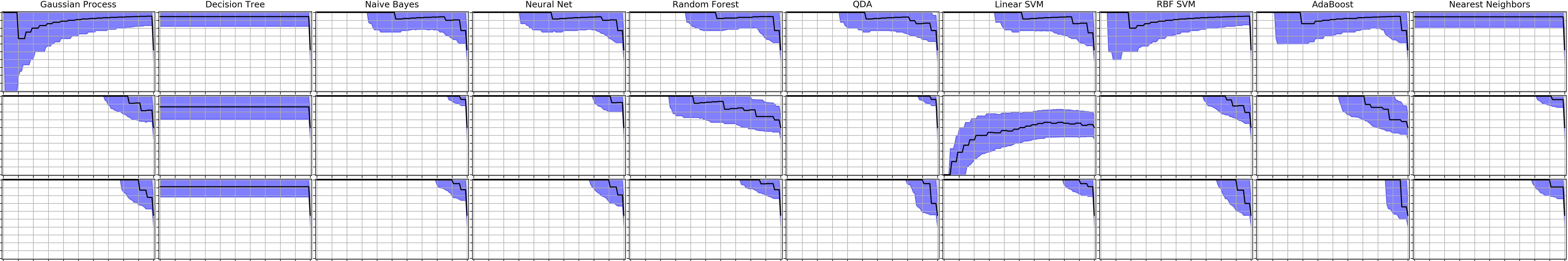
Precision-recall curves
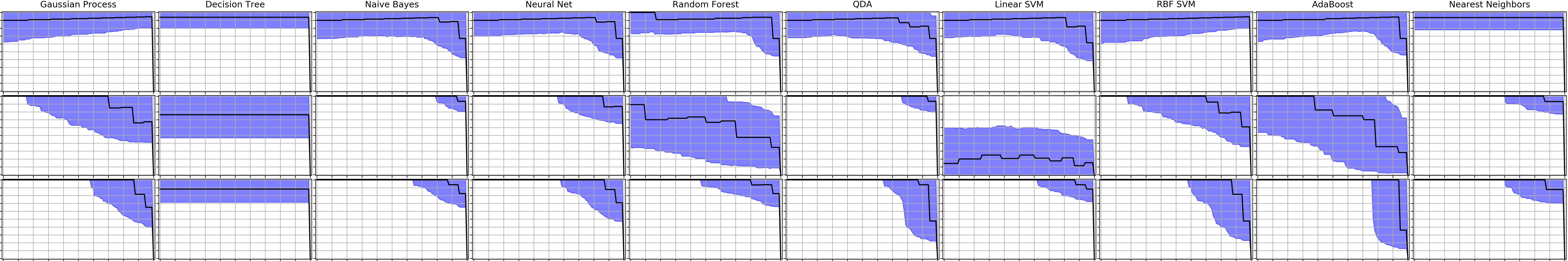
Precision-recall-gain curves
Usage for regression problems
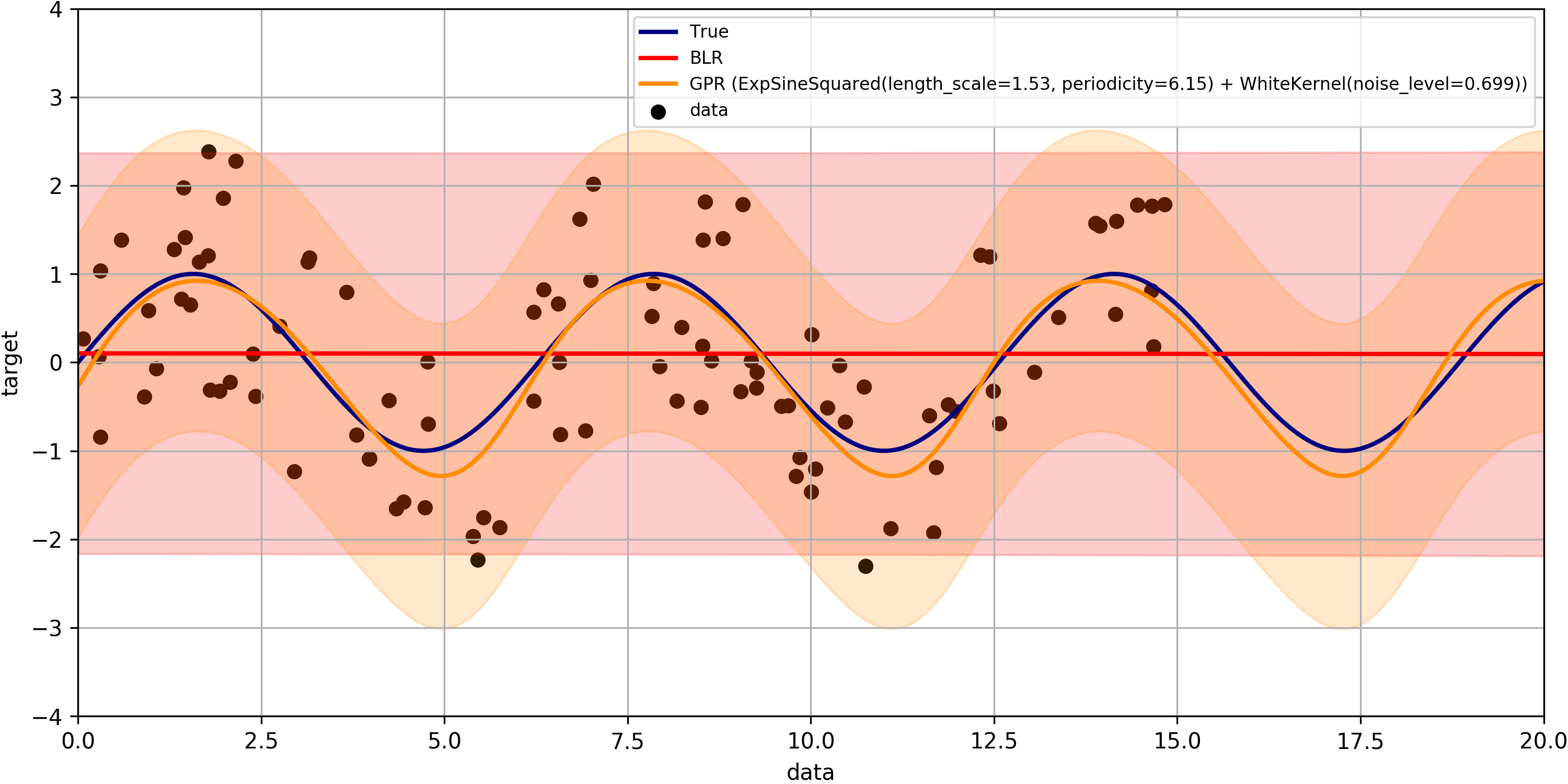
The mlpaper package can also be applied to a regression problem with:
import mlpaper.regression as btr
full_tbl = btr.just_benchmark(X_train, y_train, X_test, y_test, regressors, STD_REGR_LOSS, "iid", pairwise_CI=True)Here we have used pairwise_CI=True which makes the confidence intervals based on the uncertainty of the loss difference to the reference method rather than a confidence interval on the actual loss.
Output
By extending the sklearn regression demo we can make simple formatted tables:
MAE p MSE p NLL (nats) p BLR 0.96933(30) 0.0979 1.39881(67) 0.0665 1.58842(57) 0.9828 GPR 0.75(13) 0.0009 0.75(28) <0.0001 1.27(12) <0.0001 iid 0.96908 N/A 1.3982 N/A 1.5884 N/A
or in LaTeX:
\begin{tabular}{|l|Sr|Sr|Sr|}
\toprule
{} & {MAE} & {p} & {MSE} & {p} & {NLL (nats)} & {p} \\
\midrule
BLR & 0.96933(30) & 0.0979 & 1.39881(67) & 0.0665 & 1.58842(57) & 0.9828 \\
GPR & 0.75(13) & 0.0009 & 0.75(28) & <0.0001 & 1.27(12) & <0.0001 \\
iid & 0.96908 & N/A & 1.3982 & N/A & 1.5884 & N/A \\
\bottomrule
\end{tabular}
Installation
Only Python>=3.5 is officially supported, but older versions of Python likely work as well.
The core package itself can be installed with:
pip install mlpaperTo also get the dependencies for the demos in the README install with
pip install mlpaper[demo]Contributing
The following instructions have been tested with Python 3.7.4 on Mac OS (10.14.6).
Install in editable mode
First, define the variables for the paths we will use:
GIT=/path/to/where/you/put/repos
ENVS=/path/to/where/you/put/virtualenvsThen clone the repo in your git directory $GIT:
cd $GIT
git clone https://github.com/rdturnermtl/mlpaper.gitInside your virtual environments folder $ENVS, make the environment:
cd $ENVS
virtualenv mlpaper --python=python3.7
source $ENVS/mlpaper/bin/activateNow we can install the pip dependencies. Move back into your git directory and run
cd $GIT/mlpaper
pip install -r requirements/base.txt
pip install -e . # Install the package itselfContributor tools
First, we need to setup some needed tools:
cd $ENVS
virtualenv mlpaper_tools --python=python3.7
source $ENVS/mlpaper_tools/bin/activate
pip install -r $GIT/mlpaper/requirements/tools.txtTo install the pre-commit hooks for contributing run (in the mlpaper_tools environment):
cd $GIT/mlpaper
pre-commit installTo rebuild the requirements, we can run:
cd $GIT/mlpaper
# Check if there any discrepancies in the .in files
pipreqs mlpaper/ --diff requirements/base.in
pipreqs tests/ --diff requirements/test.in
pipreqs demos/ --diff requirements/demo.in
pipreqs docs/ --diff requirements/docs.in
# Regenerate the .txt files from .in files
pip-compile-multi --no-upgradeGenerating the documentation
First setup the environment for building with Sphinx:
cd $ENVS
virtualenv mlpaper_docs --python=python3.7
source $ENVS/mlpaper_docs/bin/activate
pip install -r $GIT/mlpaper/requirements/docs.txtThen we can do the build:
cd $GIT/mlpaper/docs
make all
open _build/html/index.htmlDocumentation will be available in all formats in Makefile. Use make html to only generate the HTML documentation.
Running the tests
The tests for this package can be run with:
cd $GIT/mlpaper
./local_test.shThe script creates an environment using the requirements found in requirements/test.txt. A code coverage report will also be produced in $GIT/mlpaper/htmlcov/index.html.
Deployment
The wheel (tar ball) for deployment as a pip installable package can be built using the script:
cd $GIT/mlpaper/
./build_wheel.shLinks
The source is hosted on GitHub.
The documentation is hosted at Read the Docs.
Installable from PyPI.
License
This project is licensed under the Apache 2 License - see the LICENSE file for details.
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
File details
Details for the file mlpaper-0.0.3.tar.gz.
File metadata
- Download URL: mlpaper-0.0.3.tar.gz
- Upload date:
- Size: 53.3 kB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/3.1.1 pkginfo/1.5.0.1 requests/2.24.0 setuptools/47.3.1 requests-toolbelt/0.9.1 tqdm/4.46.1 CPython/3.7.4
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
525add6a45c5979fbfe26172838590e66eedf6e80d8d1438102f3649e210320a
|
|
| MD5 |
cab6b337209373d2f47b1492c74187e0
|
|
| BLAKE2b-256 |
0dcbec9bea40e20dfbd335b27f11e55bc291e7f6d6dd3c477cdf2e813b698122
|











