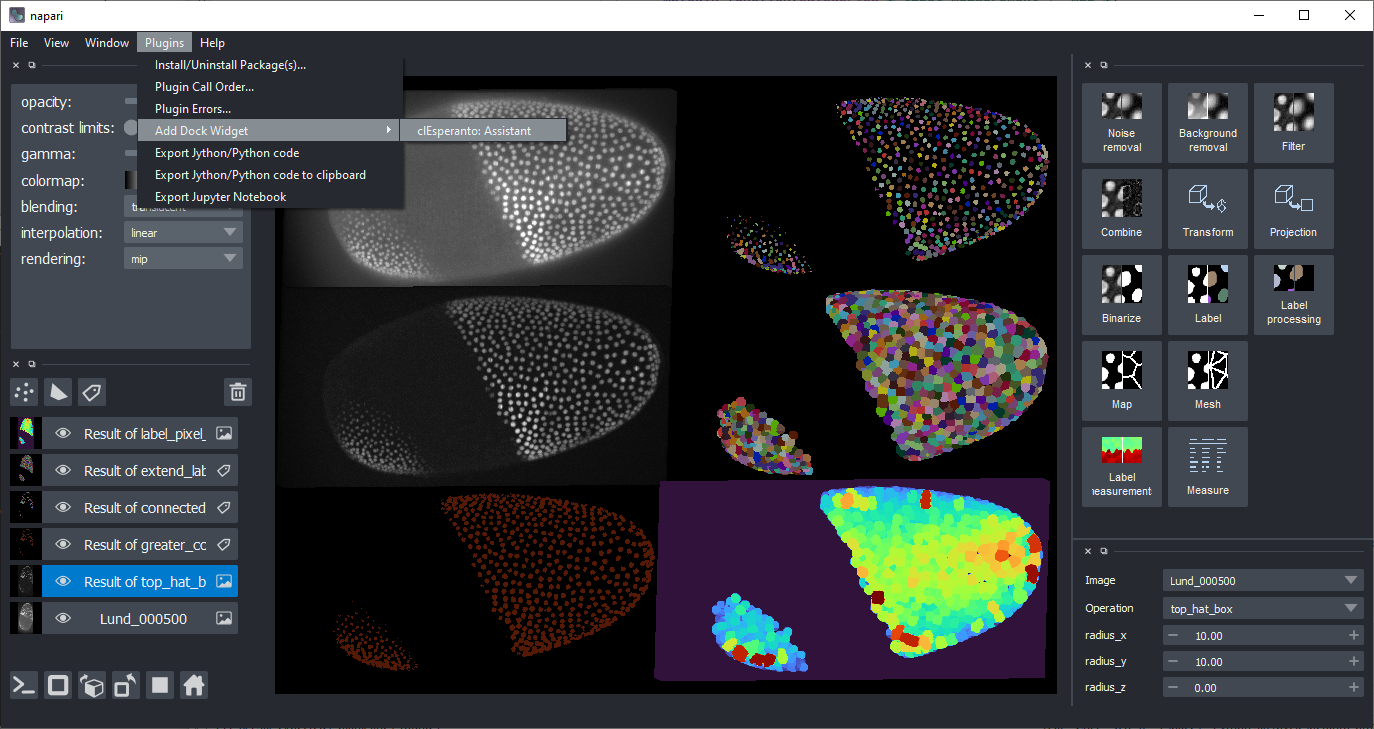
OpenCL based GPU-accelerated image processing in napari
Project description
napari-pyclesperanto-assistant
The py-clEsperanto-assistant is a yet experimental napari plugin for building GPU-accelerated image processing workflows. It is part of the clEsperanto project. It uses pyclesperanto and with that pyopencl as backend for processing images. This plugin was generated with Cookiecutter using with napari's cookiecutter-napari-plugin template.
Installation
Installation using conda
conda install -c conda-forge pyopencl
pip install napari[all] napari-pyclesperanto-assistant
Afterwards, start napari from the command line:
napari --with clEsperanto
(You can also find the assistant in the Plugins menu.)
Installation using the napari installer
Download and install napari.
Windows users please download pyopencl...cl12-cp38-cp38-win_amd64.whl. Use the command line to navigate to the folder where you downloaded it (for example the Downloads folder using cd Downloads). From there, run the following line after replacing <username> with your username:
C:\Users\<username>\AppData\Local\Programs\napari\python\python.exe -m pip install pyopencl-2020.2.2+cl12-cp38-cp38-win_amd64.whl
Start napari and navigate to its menu Plugins > Install/Uninstall Package(s).... Select napari-pyclesperanto-assistant from the list and install it by clicking the blue button on the right:
Restart napari. Afterwards, you should find the Assistant in the plugins menu:
Installation on windows
On windows some additional steps are necessary. Download a pre-compiled wheel of pyopencl e.g. from here.
It is recommended to install pyopencl-...+cl12-cp38-cp38-win_amd64 - the cl12 and cp38 are important when choosing the right download. They stand for OpenCL 1.2 and Python 3.8.
Enter the correct pyopencl filename and execute this from the command line:
pip install pyopencl-2020.3.1+cl12-cp38-cp38-win_amd64.whl
In case napari doesn't start up with an error mentioning numpy (see also), execute this from the command line:
pip install numpy==1.19.3
In case of error messages like RuntimeError: module compiled against API version 0xe but this version of numpy is 0xd try:
pip uninstall pyopencl
conda install -c conda-forge pyopencl==2021.2.1
Features
pyclesperanto offers various possibilities for processing images. It comes from developers who work in life sciences and thus, it may be focused towards processing two- and three-dimensional microscopy image data showing cells and tissues. A selection of pyclesperanto's functionality is available via the assistant user interface. Typical workflows which can be built with this assistant include
- image filtering
- denoising / noise reduction
- background subtraction
- grey value morphology
- combining images
- masking
- image math (adding, subtracting, multiplying, dividing images)
- image transformations
- translation
- rotation
- sub-stacks
- image projections
- minimum / mean / maximum / sum / standard deviation projections
- image segmentation
- binarization
- thresholding
- labeling
- regionalization
- instance segmentation
- semantic segmentation
- detect label edges
- label spots
- connected component labeling
- Voronoi-Otsu-labeling
- post-processing of binary images
- dilation
- erosion
- binary opening
- binary closing
- binary and / or / xor
- post-processing of label images
- dilation (expansion) of labels
- erosion (shrinking) of labels
- extend labels via Voronoi
- exclude labels on edges
- exclude labels within / out of size / value range
- parametric maps
- proximal / touching neighbor count
- distance measurements to touching / proximal / n-nearest neighbors
- pixel count map
- mean / maximum / extension ratio map
- label measurements / post processing of parametric maps
- minimum / mean / maximum / standard deviation intensity maps
- minimum / mean / maximum / standard deviation of touching / n-nearest / neighbors
- neighbor meshes
- touching neighbors
- n-nearest neighbors
- proximal neighbors
- distance meshes
- measurements based on label images
- bounding box 2D / 3D
- minimum / mean / maximum / sum / standard deviation intensity
- mass center
- centroid
- mean / maximum distance to centroid (and min/max ratio shape descriptor)
- mean / maximum distance to center of mass (and min/max ratio shape descriptor)
- code export
- python / Fiji-compatible jython
- python jupyter notebooks
- pyclesperanto scripting
- cell counting
- cell differentiation
- tissue classification
Usage
This short tutorial demonstrates how to generate code using the pyclersperanto-assistant.
<iframe src="docs/images/pyclesperanto_assistant_screencast.mp4" width="600" height="300"></iframe> [Download workflow as video](docs/images/pyclesperanto_assistant_screencast.mp4)Start up the assistant
Open a command line and start up the assistant and pass the image file you want to process. The shown example image can be found online
python -m napari_pyclesperanto_assistant C:\structure\code\napari_pyclesperanto_assistant\napari_pyclesperanto_assistant\data\CalibZAPWfixed_000154_max-16.tif
Alternatively, you can attach the assistant to your napari from within your python code like this:
import napari
# start napari
viewer = napari.Viewer()
viewer.window.add_plugin_dock_widget('clEsperanto')
napari.run()
napari will open with the assistant activated:
Set up a workflow
Choose categories of operations in the top right panel, for example start with denoising using a Gaussian Blur with sigma 1 in x and y:
Choose more processing steps. Note: You can change the input image/layer for each operation, the operation and its parameters in the bottom right panel. For example, continue with these steps
- Filter (Background Removal): Top hat, radius 5 in x and y
- Binarization: Threshold Otsu
- Label: Voronoi labeling
- Map: Touching neighbor count map
- Binarization: Detect label edges, with the result from the second last step as input.
Hide some layers showing intermediate results. Switch the bleinding of the final result layer to "additive" to see through it on the original image.
Code generation
In the plugins menu, you find two entries which allow you to export your workflow as Python/Jython code.
Export your workflow as Jupyter notebook. Start the notebook from the command line using
jupyter notebook my_notebook.ipynb
Alternatively, export the workflow as Jython/Python script. This script can be executed from the command line like this
python my_script.py
It can also be executed in Fiji, in case the CLIJx-assistant is installed.
Note: Depeending on which layers were visible while exporting the code, different code is exported. Only visible layers are shown. Change layer visibility and export the script again. If Fiji asks you if it should reload the script file, click on "Reload".
For developers
Getting the recent code from github and locally installing it
git clone https://github.com/clesperanto/napari_pyclesperanto_assistant.git
pip install -e ./napari_pyclesperanto_assistant
Optional: Also install pyclesperantos recent source code from github:
git clone https://github.com/clEsperanto/pyclesperanto_prototype.git
pip install -e ./pyclesperanto_prototype
Starting up napari with the pyclesperanto assistant installed:
ipython --gui=qt napari_pyclesperanto_assistant\napari_pyclesperanto_assistant
Feedback welcome!
clEsperanto is developed in the open because we believe in the open source community. See our community guidelines. Feel free to drop feedback as github issue or via image.sc
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
File details
Details for the file napari_pyclesperanto_assistant-0.9.2.tar.gz.
File metadata
- Download URL: napari_pyclesperanto_assistant-0.9.2.tar.gz
- Upload date:
- Size: 88.0 kB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/3.4.1 importlib_metadata/4.5.0 pkginfo/1.7.0 requests/2.25.1 requests-toolbelt/0.9.1 tqdm/4.61.1 CPython/3.8.5
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | b686393470f594ffa1d36acc4441ee781a36fe8bce0c8bd005a641c7197954d4 |
|
| MD5 | a6495797cc52f3a8d56fd15d40e3d3e8 |
|
| BLAKE2b-256 | 75ada5fda33b165fb4ef19648cae4327c0233f2d812f2b4cac11fabb68d03f32 |
File details
Details for the file napari_pyclesperanto_assistant-0.9.2-py3-none-any.whl.
File metadata
- Download URL: napari_pyclesperanto_assistant-0.9.2-py3-none-any.whl
- Upload date:
- Size: 91.0 kB
- Tags: Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/3.4.1 importlib_metadata/4.5.0 pkginfo/1.7.0 requests/2.25.1 requests-toolbelt/0.9.1 tqdm/4.61.1 CPython/3.8.5
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | d768f568d035b26e30a0b63e9fce39cea8351df07cd4a8a1edae6c0722efb636 |
|
| MD5 | 06219edba903629dbe5c075e87489357 |
|
| BLAKE2b-256 | 84e42f028f17e00dffa9b90b170d360c5117a7750e0b5fbeb047b8871f9f8ae4 |