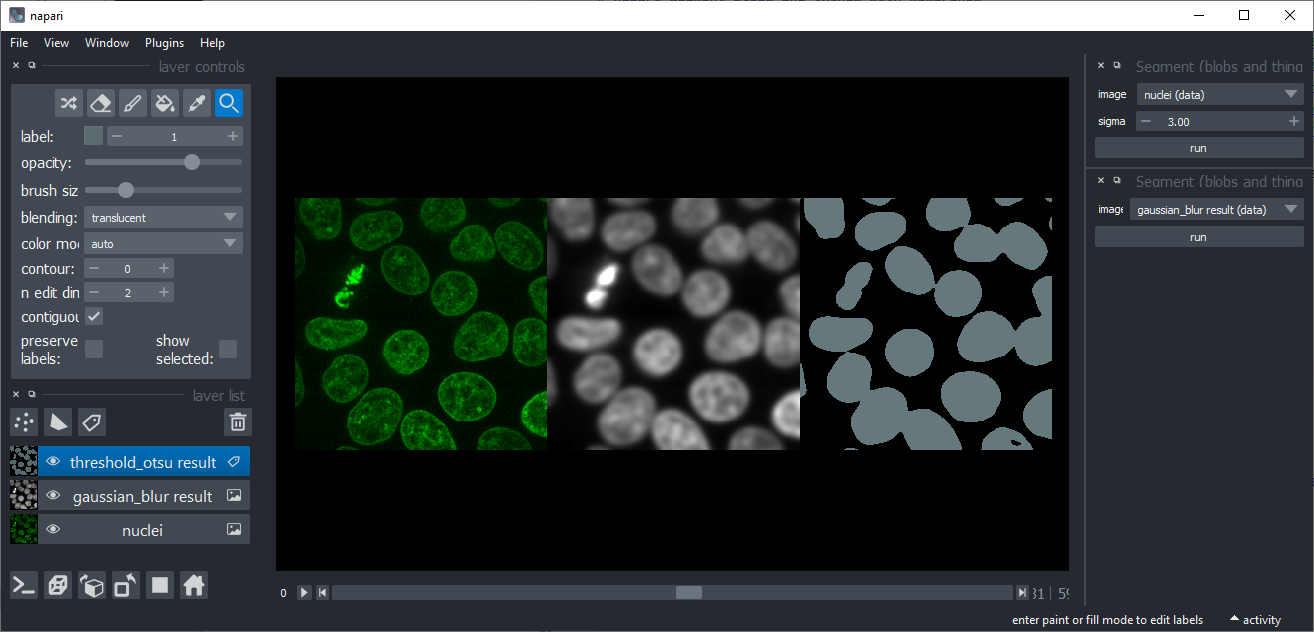
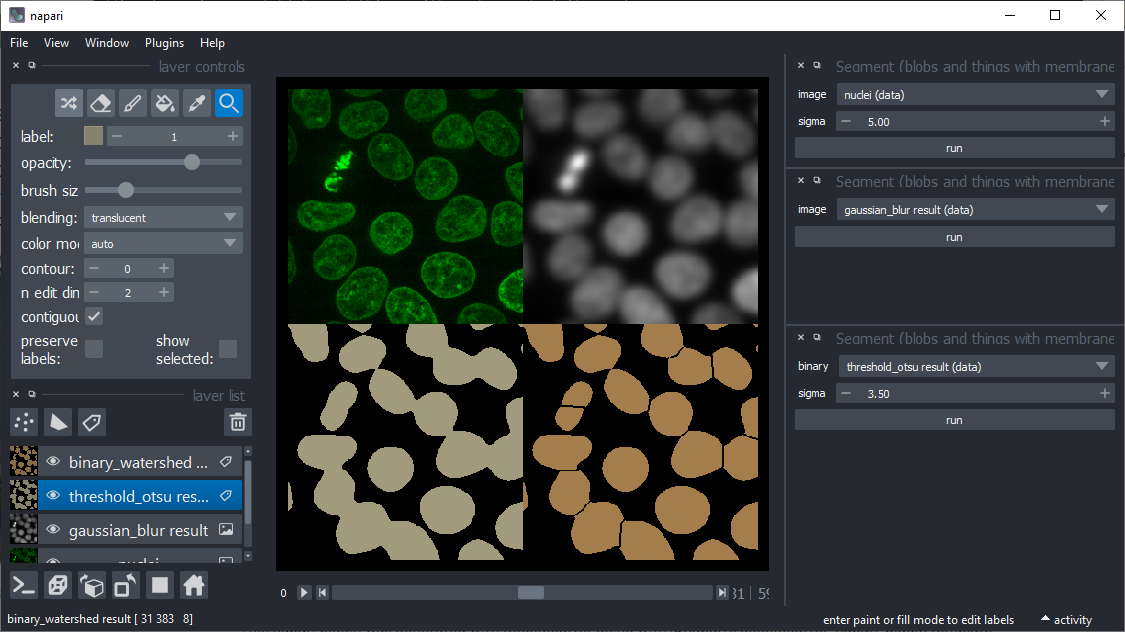
A plugin based on scikit-image for segmenting nuclei and cells based on fluorescent microscopy images with high intensity in nuclei and/or membranes
Project description
napari-segment-blobs-and-things-with-membranes
A plugin based on scikit-image for segmenting nuclei and cells based on fluorescent microscopy images with high intensity in nuclei and/or membranes. The available functions and their user interface based on magicgui are shown below. You can also call these functions as shown in the demo notebook.
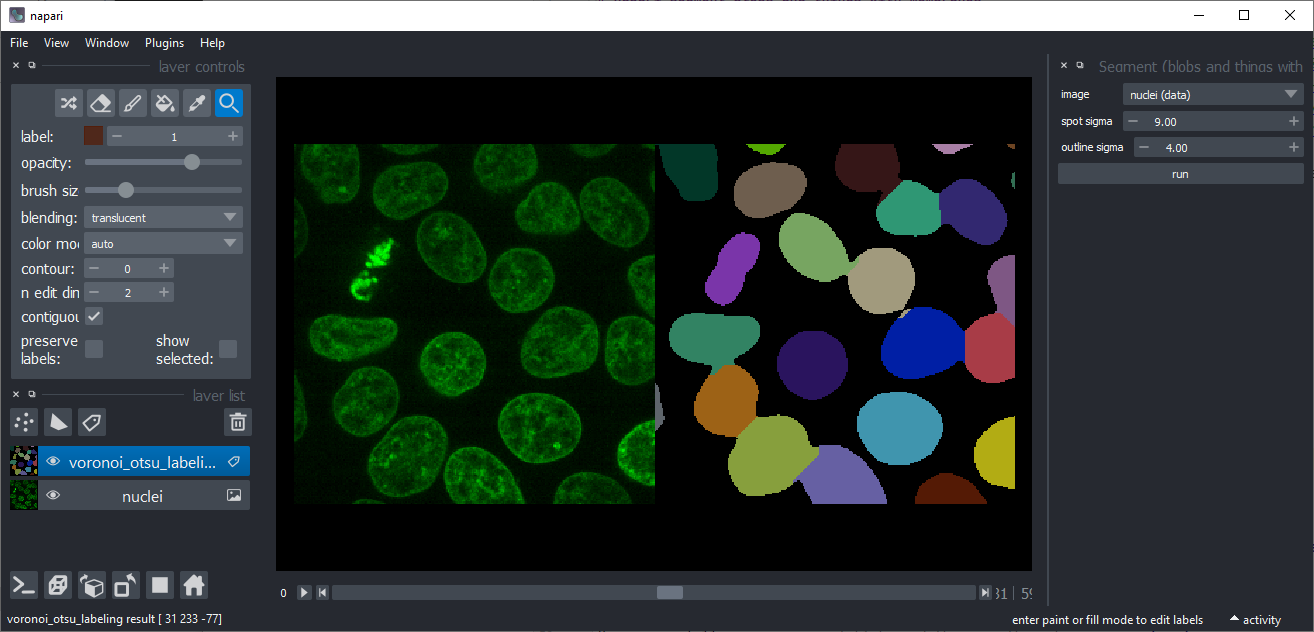
Voronoi-Otsu-Labeling
This algorithm uses Otsu's thresholding method in combination with
Gaussian blur and a
Voronoi-Tesselation
approach to label bright objects such as nuclei in an intensity image. The alogrithm has two sigma parameters which allow
you to fine-tune where objects should be cut (spot_sigma) and how smooth outlines should be (outline_sigma).
This implementation aims to be similar to Voronoi-Otsu-Labeling in clesperanto.
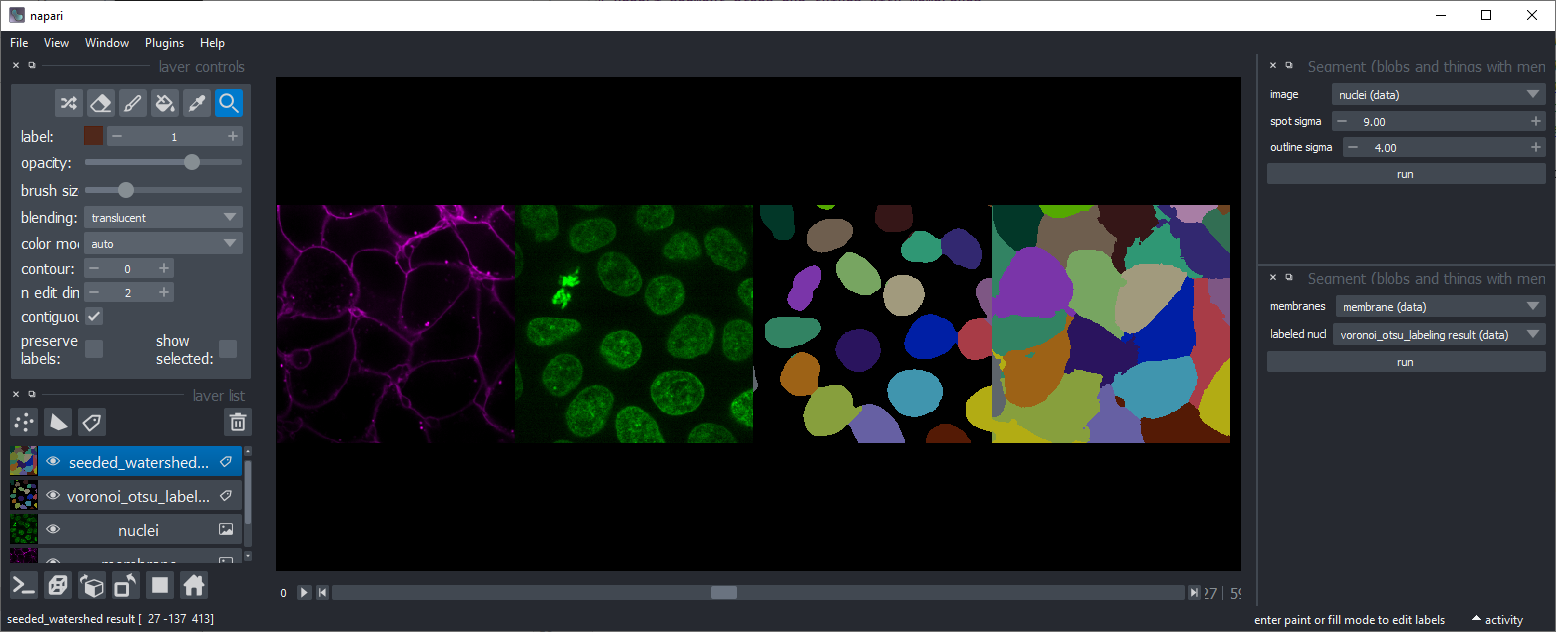
Seeded Watershed
Starting from an image showing high-intensity membranes and a seed-image where objects have been labeled (e.g. using Voronoi-Otsu-Labeling), objects are labeled that are constrained by the membranes.
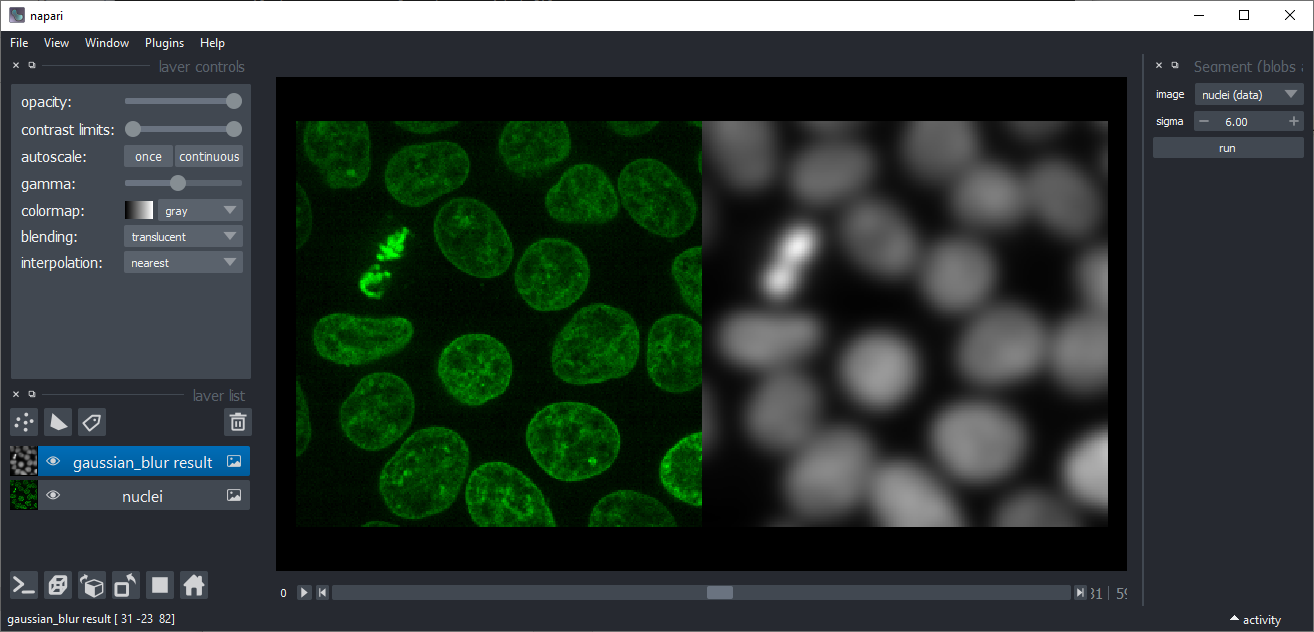
Gaussian blur
Applies a Gaussian blur to an image. This might be useful for denoising, e.g. before applying the Threshold-Otsu method.
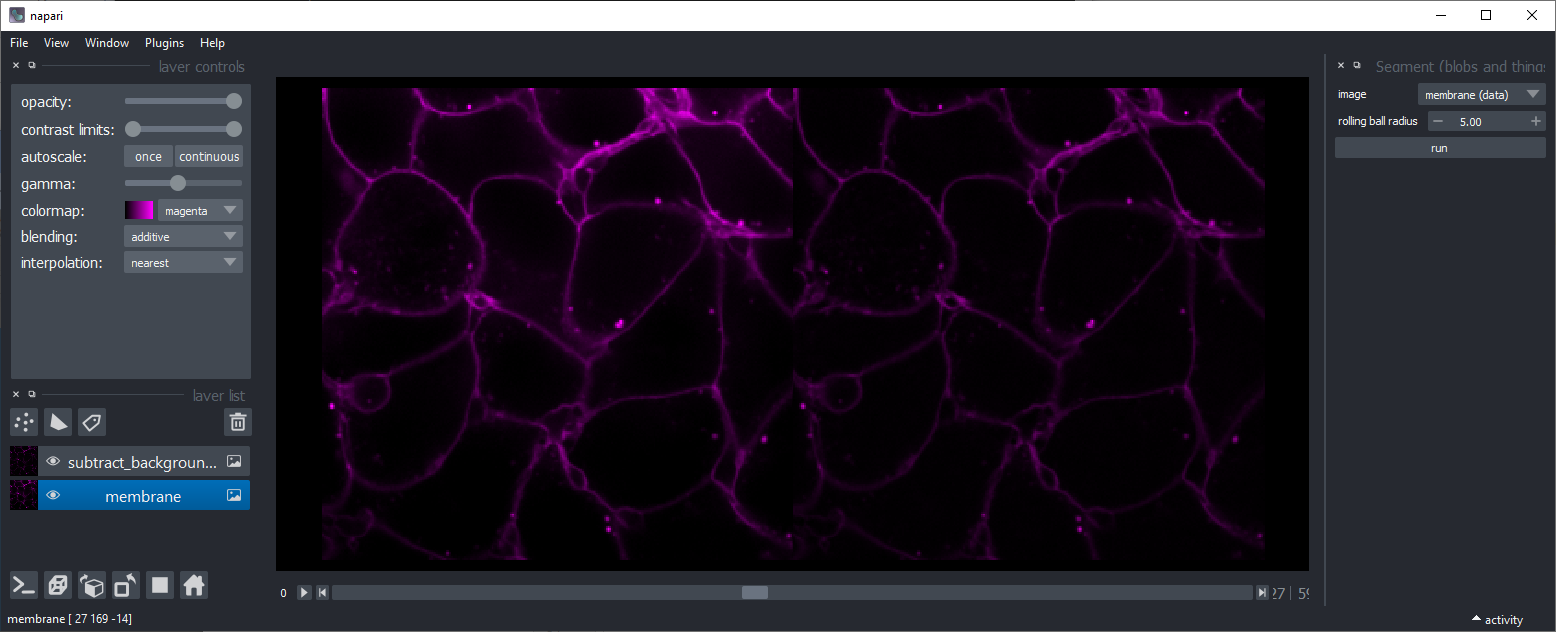
Subtract background
Subtracts background using scikit-image's rolling-ball algorithm. This might be useful, for example to make intensity of membranes more similar in different regions of an image.
Threshold Otsu
Binarizes an image using scikit-image's threshold Otsu algorithm, also known as Otsu's method.
Binary watershed
In case objects stick together after thresholding, this implementation of the watershed algorithm might help. It aims to deliver similar results as ImageJ's watershed implementation. This function appears to work well in 2D only at the moment.
Connected components labeling
Takes a binary image and produces a label image with all separated objects labeled differently. Under the hood, it uses scikit-image's label function.
This napari plugin was generated with Cookiecutter using with @napari's cookiecutter-napari-plugin template.
Installation
You can install napari-segment-blobs-and-things-with-membranes via pip:
pip install napari-segment-blobs-and-things-with-membranes
Contributing
Contributions are very welcome. Tests can be run with tox, please ensure the coverage at least stays the same before you submit a pull request.
License
Distributed under the terms of the BSD-3 license, "napari-segment-blobs-and-things-with-membranes" is free and open source software
Issues
If you encounter any problems, please create a thread on image.sc along with a detailed description and tag @haesleinhuepf.
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
Hashes for napari-segment-blobs-and-things-with-membranes-0.1.1.tar.gz
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 0f5fe7b1928544b0bd6cea4786438d2215f06d33663ea7439a7ad3824f43db33 |
|
| MD5 | 54940fc0542b16115ed7b8301b5112bc |
|
| BLAKE2b-256 | 723d71735b95101a3d5b38d93838e28c27af8b920ca98265bfd055c9ccdfac5b |
Hashes for napari_segment_blobs_and_things_with_membranes-0.1.1-py3-none-any.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 2b283591b81381a3a45576f6d3dc74c37a356523009598636bfecbe2c03f6e6e |
|
| MD5 | af025cb1bb17175cb8394f6d53013160 |
|
| BLAKE2b-256 | 05bf7a26329db1014f8fb673ff61bd9ce9ec45623828fdb28830ee0a40b46114 |