No project description provided
Project description
nnx
Nerual Networks for JAX
nnx is a lightweight module system for JAX that provides the same power as flax but with a simpler mental model and implementation closer to equinox. It is built on top of refx, which enables shared state, tractable mutability, and semantic partitioning. nnx also supports stateful transformations, allowing you to train your models efficiently.
Status
nnx is currently a proof of concept, it is meant to explore the design space of a lightweight module system for JAX based on Refx.
Installation
To get started with nnx, first install the package using pip:
pip install nnx
Usage
import nnx
import jax
class Linear(nnx.Module):
w: jax.Array = nnx.param()
b: jax.Array = nnx.param()
def __init__(self, din: int, dout: int):
key = self.make_rng("params")
self.w = jax.random.uniform(key, (din, dout))
self.b = jax.numpy.zeros((dout,))
def __call__(self, x):
return x @ self.w + self.b
model = Linear.init(jax.random.PRNGKey(0))(din=12, dout=2)
@nnx.jit
def train_step(model, x, y):
def loss_fn(model):
y_pred = model(x)
return jax.numpy.mean((y_pred - y) ** 2)
grad = nnx.grad(loss_fn, wrt="params")(model)
model["params"] = jax.tree_map(lambda w, g: w - 0.1 * g, model["params"], grad)
# yes... there's no return :)
train_step(model, x, y)
Design
Modules
NNX Modules are simple_pytree Pytrees with a few additional features to make them more easier to use with refx references. A custom Module can be created simply by
subclassing nnx.Module and marking which fields are references with nnx.ref or nnx.param, as we will explain later, these are descriptors that store a Ref instance in a separate attribute and make using references transparent to the user.
Here is an example of a simple Linear module:
import nnx
import jax
class Linear(nnx.Module):
w: jax.Array = nnx.ref("params")
b: jax.Array = nnx.param() # shortcut for ref("params")
def __init__(self, din: int, dout: int):
key = self.make_rng("params") # request an RNG key
self.w = jax.random.uniform(key, (din, dout))
self.b = jax.numpy.zeros((dout,))
def __call__(self, x):
return x @ self.w + self.b
NNX offers the same make_rng API as Flax to distribute RNG keys where they are needed, it does this by storing RNG keys in a global state and carefully handling them with context managers. You can set the state for the RNG keys and other flags via the init and apply methods, which are similar to Flax's init and apply methods but designed to be friendlier with static analysis tools.
# global state ==> .....................
model = Linear.init(jax.random.PRNGKey(0))(din=12, dout=2)
# constructor args ==> ^^^^^^^^^^^^^^^^
If global state is not needed you can just use the constructor directly.
RefField Descriptor
nnx.ref and nnx.param are descriptors that create RefField instances. RefField is a descriptor that stores a Ref instance in a separate {attribute_name}__ref attribute, and handle retrieving and setting the value of the reference automatically so that the user doesn't have to manipulate references directly. RefField inherits from dataclasses.Field in order to be compatible with dataclasses when needed.
Here is a simplified version of how RefField is implemented:
class RefField(dataclasses.Field):
def __set_name__(self, cls, name):
self.name = name
def __get__(self, obj, objtype=None):
ref = getattr(obj, f"{self.name}__ref")
return ref.value
def __set__(self, obj, value):
ref = getattr(obj, f"{self.name}__ref")
ref.value = value
The only thing to note here is that Refs are created during the first call to __set__ if the companion {name}__ref attribute doesn't exist yet. This should only happen during __init__ or else Module will raise an error as simple_pytree Pytrees are frozen after initialization.
GetItem and SetItem syntactic sugar
Module implements __getitem__ and __setitem__ to provide syntactic sugar for creating and updating Partitions. Despite the appearance, __setitem__ does not modify the Module's structure, it just updates the values of the references as can be seen in this simplified implementation:
class Module(simple_pytree.Pytree):
...
def __getitem__(self, collection: str) -> refx.Partition:
derefed_module = refx.deref(self)[0]
return nnx.get_partition(derefed_module, collection)
def __setitem__(self, collection: str, updates: refx.Partition):
partition = nnx.get_partition(self, collection)
refx.update_refs(partition, updates)
Sample usage could look something like this:
model["params"] = jax.tree_map(lambda w, g: w - 0.1 * g, model["params"], grad)
Here model["params"] is a Partition that contains all the references in the params collection, and grad is a Partition with the same structure as model["params"] but with gradients instead of parameters. moduel["params"] = ... updates the values of the references in model["params"] with the values of the sdg update.
Transformations
Currently NNX offers 3 types of transformations: stateful, filtered, and the partition API. At the moment its not clear which API is the best, the 3 will be kept for now.
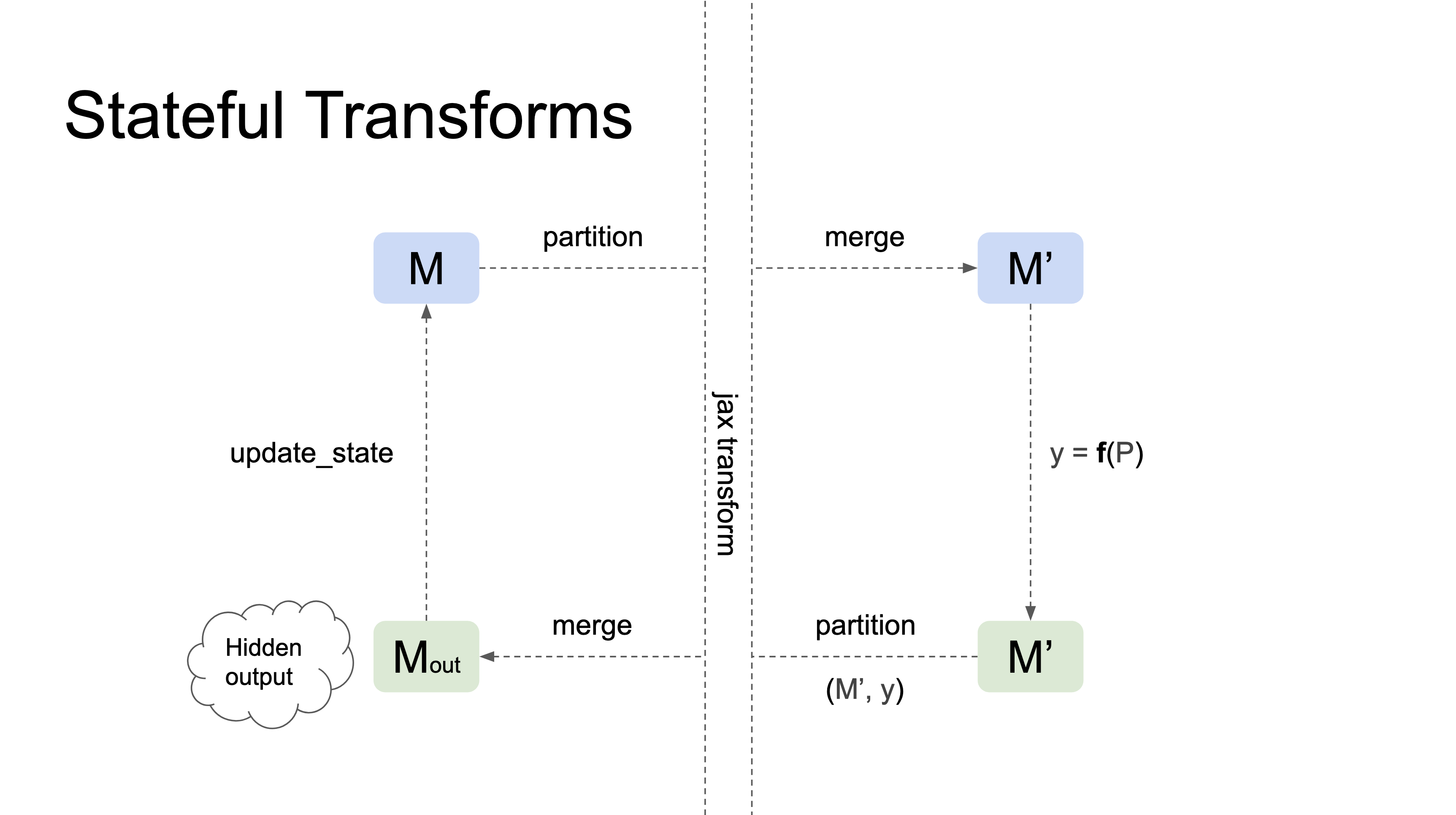
Stateful Transforms
Stateful Transforms take a pytree of references (e.g. a Module) as their first argument, track changes in the state of the references that happen inside the transformation, and automatically propagate those changes to the input pytree outside the transformation. In general they have the following properties:
- they behave as stateful functions w.r.t. the first argument
- they can operate on collections and RNG streams according to the transformation's semantics, exactly like Flax's transformations
- they take care of handling all relevant global state such as
nnx.scopeand Refx's trace state
Here is a diagram of how stateful transformations work:
Currently nnx.jit and nnx.grad are the only stateful transformations.
Here is how a train_step function could be implemented using nnx.jit and nnx.grad:
@nnx.jit
def train_step(model, x, y):
def loss_fn(model):
y_pred = model(x)
return jax.numpy.mean((y_pred - y) ** 2)
# compute gradient
grad = nnx.grad(loss_fn, wrt="params")(model)
# sdg update
model["params"] = jax.tree_map(lambda w, g: w - 0.1 * g, model["params"], grad)
# stateful update, no return !!!
train_step(model, x, y)
The most interesting part of the design is that the code looks very imperative since the state is automatically propagated in and out of the transformations. However, more thought is needed to see how to correctly support jit's in_shardings and out_shardings arguments.
Filtered Transformations
Filtered transformations are more flexible in that they can take pytrees of references in any of their arguments and also return pytrees of references. They just deref and reref all their inputs and outputs to move the pytrees across the transformation. In general they have the following properties:
- they behave as pure functions
- they don't handle any global state except for Refx's trace state
Currently nnx.jit_filter is the only filtered transformation.
Here is how a train_step function could be implemented using nnx.jit_filter and nnx.grad:
@nnx.jit_filter
def train_step(model, x, y):
def loss_fn(model):
y_pred = model(x)
return jax.numpy.mean((y_pred - y) ** 2)
# compute gradient
grad = nnx.grad(loss_fn, wrt="params")(model)
# sdg update |--------sdg-----------|
model["params"] = jax.tree_map(lambda w, g: w - 0.1 * g, model["params"], grad)
return model
model = train_step(model, x, y)
Filtered transformations must output any state that they want to propagate but have more flexibility in how they handle it. Adding support for jit's in_shardings and out_shardings arguments is probably more straightforward than with stateful transformations.
Partition API
The partition API mimicks Flax's variables plus apply API. It splits a pytree of references into all its Partitions and creates a ModuleDef object that knows how to reconstruct the original pytree from the Partitions. Since each Partition is a flat dictionary, this API works with regular JAX transformations.
Here is a diagram of how the partition API works:
Here is an example of how to use the partition API:
model: ModuleDef
partitions, model = model.partition()
params = partitions["params"]
@jax.jit
def train_step(params, x, y):
def loss_fn(params): # |----merge----|
y_pred, updates = model.apply([params])(x)
return jax.numpy.mean((y_pred - y) ** 2)
# compute gradient
grad = jax.grad(loss_fn)(params)
# sdg update |--------sdg-----------|
params = jax.tree_map(lambda w, g: w - 0.1 * g, params, grad)
return params
params = train_step(params, x, y)
The main benefit of the partition API is that its more compatible with other JAX tools as the training step can be written using regular JAX transformations. The main drawback is that it's more verbose and users have to manually keep track of all the partitions, this overhead is often what makes flax and haiku a bit harder to learn than other frameworks like pytorch and keras.
Case Studies
Shared State
In nnx, it's possible to create modules that share state between each other. This can be useful when designing complex neural network architectures, as it allows you to reuse certain layers and reduce the number of learnable parameters.
Here's an example of how to create a module with shared state:
class Block(nnx.Module):
def __init__(self, linear: nnx.Linear):
self.linear = linear
self.bn = nnx.BatchNorm(2)
def __call__(self, x):
return nnx.relu(self.bn(self.linear(x)))
class Model(nnx.Module):
def __init__(self):
shared = nnx.Linear(2, 2)
self.block1 = Block(shared)
self.block2 = Block(shared)
def __call__(self, x):
x = self.block1(x)
x = self.block2(x)
return x
In this example, the Model module contains two instances of the Block module, each of which shares the same nnx.Linear module. To run the model you can use the apply method to set the use_running_average flag for all BatchNorm modules.
Here's an example of how to compute the loss for a Model instance:
def loss_fn(model: Model, x: jax.Array, y: jax.Array):
y_pred = model.apply(use_running_average=False)(x)
return jnp.mean((y - y_pred) ** 2)
It's worth noting that the state for the shared nnx.Linear module will be kept in sync at all times on both Block instances, including during gradient updates.
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
Filter files by name, interpreter, ABI, and platform.
If you're not sure about the file name format, learn more about wheel file names.
Copy a direct link to the current filters
File details
Details for the file nnx-0.0.2.tar.gz.
File metadata
- Download URL: nnx-0.0.2.tar.gz
- Upload date:
- Size: 25.8 kB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: poetry/1.4.0 CPython/3.8.10 Linux/5.13.0-1027-gcp
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
bf07381924909b5f59fd5f1d4fd16451c620982364f9427fa5c24ad0efdb64be
|
|
| MD5 |
175909785197c5f6c96499ca8bf8130f
|
|
| BLAKE2b-256 |
5ed16b611dce750c704aa14d765896e83a430ae377b34be0683bb03f9ccb2c8b
|
File details
Details for the file nnx-0.0.2-py3-none-any.whl.
File metadata
- Download URL: nnx-0.0.2-py3-none-any.whl
- Upload date:
- Size: 26.6 kB
- Tags: Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: poetry/1.4.0 CPython/3.8.10 Linux/5.13.0-1027-gcp
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
af850627c4dd06823ba2b62ba2f99232e7897990cdf31661e31b4dee038bac32
|
|
| MD5 |
a3105eb81e0cd967a5ddbaf3412e8082
|
|
| BLAKE2b-256 |
dbc18ef078a5c9eb687811fec1ae30c54afba818029b9095222290b8f8d5701f
|