No project description provided
Project description
NNX
Neural Networks for JAX
NNX is a Neural Networks library for JAX that provides a simple yet powerful module system that adheres to standard Python semantics. Its aim is to combine the robustness of Flax with a simplified, Pythonic API akin to that of PyTorch.
- Pythonic: Modules are just regular python classes, they contain their own state, are fully mutable, and allow sharing references between Modules.
- Fully Compatible: Easy convert back and forth between Modules and pure pytrees compatible with any JAX transformation and other JAX tooling.
- Safe: NNX incorporates mechanisms to try to prevent tracer leakage, avoid stale RNGs, and ensure proper state propagation in order to help produce correct JAX programs.
- Semantic: Partition a Module's state into different semantic collections, allowing for fine-grained control when applying JAX transformations.
- Lifted Transforms: NNX offers a set of Module-aware transforms that automatically manage the Module's state and provide APIs to instruct the underlying JAX transform how to handle each state collection.
Table of Contents
Installation
To get started with nnx, install the package via pip:
pip install nnx
For the most recent version, install directly from our GitHub repository:
pip install git+https://github.com/cgarciae/nnx
Getting Started
The following example guides you through creating a basic Linear model with NNX and executing a forward pass. It also demonstrate how handle mutable state by showing how to keep track of the number of times the model has been called.
import nnx
import jax
import jax.numpy as jnp
class Linear(nnx.Module):
def __init__(self, din: int, dout: int, *, ctx: nnx.Context):
key = ctx.make_rng("params")
self.w = nnx.param(jax.random.uniform(key, (din, dout)))
self.b = nnx.param(jnp.zeros((dout,)))
self.count = nnx.var("counts", 0) # track the number of calls
def __call__(self, x):
self.count += 1
return x @ self.w + self.b
ctx = nnx.Context(params=jax.random.PRNGKey(0))
model = Linear(din=12, dout=2, ctx=ctx)
# Forward pass and verify the call count
x = jnp.ones((8, 12))
y = model(x)
assert model.count == 1
Training
Here we show case two different approaches to training the model defined in the previous secition. The first one uses lifted transforms and the second one uses the functional API. Both approaches are equivalent and produce the same results.
Lifted Transforms
In this example, we uset the nnx.jit and nnx.grad lifted transforms to define the training step. The model is trained using Stochastic Gradient Descent (SGD) and train_step doesn't require a return statement in this case as the model's state is automatically updated.
@nnx.jit
def train_step(model, x, y):
def loss_fn(model):
y_pred = model(x)
return jax.numpy.mean((y_pred - y) ** 2)
# compute gradient
grads: nnx.State = nnx.grad(loss_fn, wrt="params")(model)
# SGD update
model.update_state(
jax.tree_map(lambda w, g: w - 0.1 * g, model.filter("params"), grads)
)
# execute the training step
train_step(model, x, y)
assert model.count == 2
Note: Using nnx.jit can introduce some overhead when compared to using jax.jit directly. Use nnx.jit for simple prototypes and getting started quickly. For more advanced use cases, use the functional API.
Functional API
In this example, we utilize the functional API for training. This approach provides more control over the state and allows you to use regular JAX transformations. The model is also trained using Stochastic Gradient Descent (SGD).
(params, counts), moduledef = model.partition("params", "counts")
@jax.jit
def train_step(params, counts, x, y):
def loss_fn(params):
y_pred, (updates, _) = moduledef.apply(params, counts)(x)
loss = jax.numpy.mean((y_pred - y) ** 2)
return loss, updates.filter("counts")
# compute gradient
grads, counts = jax.grad(loss_fn, has_aux=True)(params)
# SGD update
params = jax.tree_map(lambda w, g: w - 0.1 * g, params, grads)
return params, counts
# execute the training step
params, counts = train_step(params, counts, x, y)
model = moduledef.merge(params, counts)
assert model.count == 2
Examples
- Using Lifted Transforms: Shows how to train a simple model using lifted transforms.
- Using the Functional API: Shows how to train a simple model using the functional API.
- Using TrainState: Shows how to train a simple model using the functional API with the help of
TrainState. - Using PureModule (experimental): Shows how to train a simple model using the functional API and leveraging
PureModuleto simplify the code. - Training a VAE: Shows how to train a VAE on the binarized MNIST dataset, uses the functional API,
TrainState, and shows how to use capture intermediate values to retrievekl_loss. - Scan over layers: An contrived example that implements scan over layers with dropout and a share BatcNorm layer to showcase how lifted transforms can be implemented. It uses the functional API along with
jax.vmapandjax.lax.scan. - Creating a Transformer: Shows how to create a Transformer with an auto-regressive decoder that uses scan over layers and a kv-cache for fast inference. Credits to @levskaya.
FAQs
Status
NNX is still in early development so expect bugs and breaking changes. That said, current API is the result of months of experimentation and we don't expect any major changes in the near future.
How is it different from Flax?
NNX takes the best features that allow Flax to scale to large projects and integrates them into a much simpler Module system with pythonic semantics.
One place in which NNX strongly deviates from Flax is that (currently) it avoids shape inference in favor of static initialization. It is not a technical limitation but rather a design choice. This design both simplifies the internal implementation and makes it easier to reason about the code for the user, at the cost of being more verbose at times. On the other hand, Pytorch users will feel right at home.
How is it different from Equinox?
While they might look similar at a surface-level, NNX's Module system is more powerful and flexible than Equinox's, it contains the following additional features:
- Uses regular python classes (no mandatory dataclass behavior).
- Modules are mutable
- Reference sharing between Modules is allowed
- Mutable state lives inside the Module (no need for a separate State container).
- Supports node metadata and semantic partitioning.
One major difference between the two frameworks is that, by design, NNX Modules are not Pytrees. This adds a safety layer as it prevents state updates from being lost by accident due to referential transparency. It also removes the need of threading a separate State container throughout the code in order to propagate state. In NNX state updates are either always preserved or explicitly discarded by the user.
User Guide
Modules
NNX Modules are normal python classes, they obey regular python semantics such as mutability and reference sharing, including reference cycles. They can contain 2 types of attributes: node attributes and static attributes. Node attributes include NNX Variables (e.g. nnx.param), Numpy arrays, JAX arrays, submodules Modules, and other NNX types. All other types are treated as static attributes.
class Foo(nnx.Module):
def __init__(self, ctx: nnx.Context):
# node attributes
self.variable = nnx.param(jnp.array(1))
self.np_buffer = np.array(2)
self.jax_buffer = jnp.array(3)
self.node = nnx.node([4, 5])
self.submodule = nnx.Linear(2, 4, ctx=ctx)
# static attributes
self.int = 1
self.float = 2.0
self.str = "hello"
self.list = [1, 2, 3]
ctx = nnx.Context(jax.random.PRNGKey(0))
model = Foo(din=12, dout=2, ctx=ctx)
As shown above, python container types such as list, tuple, and dict are treated as static attributes, if similar functionality is needed, NNX provides the Sequence and Map Modules.
Functional API
NNX Modules are not pytrees so they cannot be passed to JAX transformations. In order to interact with JAX, a Module must be partitioned into a State and ModuleDef objects. The State object is a flat dictionary-like pytree structure that contains all the deduplicated node attributes, and the ModuleDef contains the static attributes and structural information needed to reconstruct the Module.
state, moduledef = model.partition()
State({
('jax_buffer',): Array(3),
('node',): Node(value=[4, 5]),
('np_buffer',): array(2),
('submodule', 'bias'): Variable(collection='params', value=Array(...)),
('submodule', 'kernel'): Variable(collection='params', value=Array(...)),
('variable',): Variable(collection='params', value=Array(1))
})
State and ModuleDef are pytrees so they can be passed to JAX transformations. More over, ModuleDef provides 2 very important methods: merge and apply. The merge method can be used to create a new Module from a State object:
model = moduledef.merge(state)
This can be use to e.g. recreate a module inside a JAX transformation. The apply provides a functional interface to the module, it can be used call any method or submodule and get the output and the updated state:
# run __call__
y, (state, moduledef) = moduledef.apply(state)(x)
# run some_method
y, (state, moduledef) = moduledef.apply(state).some_method(x)
# run submodule
y, (state, moduledef) = moduledef.apply(state).submodule(x)
apply can call any nested method or submodule as long as it can be accessed via the . or [] operators.
Partitioning State
nnx.var lets you create Variable attributes with collection metadata, this metadata can be used to partition the state into multiple substates. nnx.param is a special case of nnx.var where collection="params".
Here are various examples of how you can use the partition method along with collection names to partition a module into multiple substates:
# partition the module into the state with all the nodes and the moduledef
state, moduledef = model.partition()
# verify that the state contains only params, else raise an error
params, moduledef = model.partition("params")
# split the state into params and batch_stats, verify no nodes are left
(params, batch_stats), moduledef = model.partition("params", "batch_stats")
# if there are any nodes left, use the `...` filter to capture them
(params, batch_stats, rest), moduledef = model.partition("params", "batch_stats", ...)
# using `...` as the only filter is equivalent to not passing any filters
model.partition(...) = model.partition()
partition will make sure all nodes are match by atleast one filter, else it will raise an error. If you have non-Variable nodes like nnx.node, jax.Array, or numpy.ndarray attributes, you can use the ... filter which will match any node. For a more general filter you can pass a predicate function of the form:
(path: Tuple[str, ...], value: Any) -> bool
To reconstruct the module from a set of substates, you can use merge as usual but passing the substates as additional arguments:
model = moduledef.merge(params, batch_stats, rest)
The same is true for apply.
y, (state, moduledef) = moduledef.apply(params, batch_stats, rest)(x)
Note that apply will return a single state object, if you need to re-partition the state you can use State's own partition method:
params, batch_stats, rest = state.partition("params", "batch_stats", ...)
Alternatively, if you are just interested in a subset of partitions, you can use the State.filter method which will not raise an error if some nodes are not matched by any filter:
# only get params
params = state.filter("params")
# get params and batch_stats
params, batch_stats = state.filter("params", "batch_stats")
Capturing Intermediate Values
In NNX you can easily propagate intemediate values by simply assigning them to an attribute at runtime. For convenience, you should assign them to a Variable attribute with a collection name by using nnx.var so you can easily retrieve them later.
Here is an example of how to create a Linear module that captures its output into a Variable attribute with the intermediates collection name:
class Linear(nnx.Module):
def __init__(self, din: int, dout: int, *, ctx: nnx.Context):
key = ctx.make_rng("params")
self.w = nnx.param(jax.random.uniform(key, (din, dout)))
self.b = nnx.param(jnp.zeros((dout,)))
def __call__(self, x):
y = x @ self.w + self.b
self.y = nnx.var("intermediates", y)
return y
ctx = nnx.Context(jax.random.PRNGKey(0))
model = Linear(12, 2, ctx=ctx)
Since y is only created when the module is called, it is not available upon initialization. However, once you call the module y will be created. It is recommended that you use pop_state to retrieve temporary collections like intermediates:
y = model(jnp.ones((8, 12)))
intermediates = model.pop_state("intermediates")
pop_state will return a State object with the nodes that match the given filter and remove them from the module's attributes.
State({
('y',): Variable(
collection='intermediates',
value=Array(...)
)
})
If you use the functional API to call the module instead, the intermediates nodes will be present in the output state. To retrieve the intermediates nodes and optionally separate them from the output state you can use State.partition:
state, moduledef = model.partition()
y, (state, moduledef) = moduledef.apply(state)(jnp.ones((8, 12)))
# "pop" the intermediates from the state
intermediates, state = state.partition("intermediates", ...)
Alternatively, you can use State.filter to retrieve the intermediates nodes without removing them from the state.
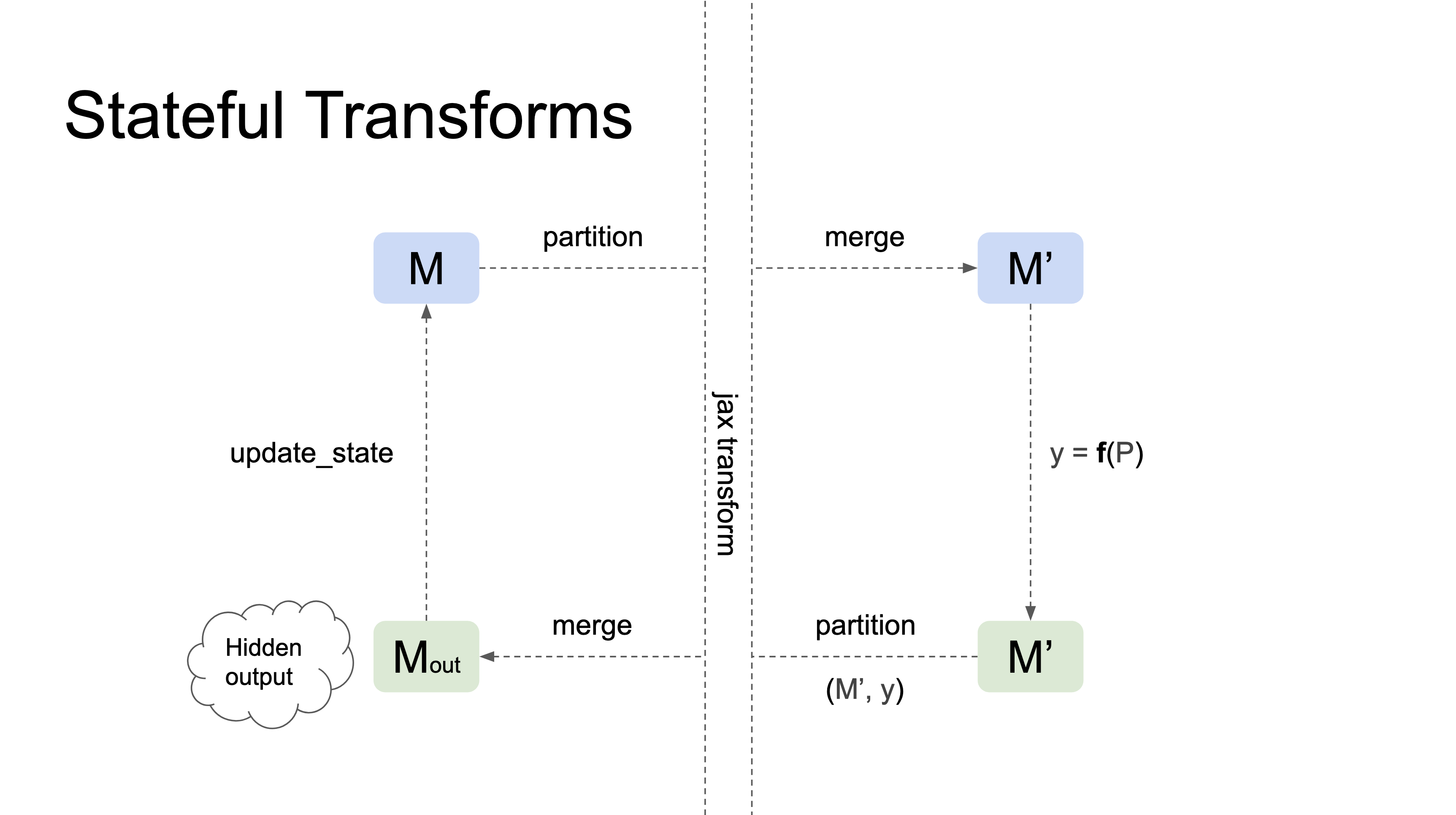
Stateful Transforms
Stateful transforms take a Module as their first argument, track changes in the state that occur within the transformation, and automatically propagate those changes to the input Module outside the transformation. In general, they behave as stateful functions with respect to the first argument.
Here's a diagram illustrating how stateful transformations work:
Currently, nnx.jit and nnx.grad are the only stateful transformations.
The following example demonstrates how a train_step function can be implemented using nnx.jit and nnx.grad:
@nnx.jit
def train_step(model, x, y):
def loss_fn(model):
y_pred = model(x)
return jax.numpy.mean((y_pred - y) ** 2)
# compute gradient
grads: nnx.State = nnx.grad(loss_fn, wrt="params")(model)
# SGD update
model.update_state(
jax.tree_map(lambda w, g: w - 0.1 * g, model.filter("params"), grads)
)
# stateful update
train_step(model, x, y)
The most interesting aspect of this design is that the code appears very imperative, as the state is automatically propagated in and out of the transformations.
Case Studies
Shared State
In NNX, you can create modules that share state between them. This is useful when designing complex neural network architectures, as it allows you to reuse certain layers and reduce the number of learnable parameters.
Here's an example of creating a module with shared state:
class Block(nnx.Module):
def __init__(self, linear: nnx.Linear, *, ctx: nnx.Context):
self.linear = linear
self.bn = nnx.BatchNorm(2, ctx=ctx)
def __call__(self, x, *, ctx: nnx.Context):
x = self.linear(x)
x = self.bn(x, ctx=ctx)
x = nnx.relu(x)
return x
class Model(nnx.Module):
def __init__(self, *, ctx: nnx.Context):
shared = nnx.Linear(2, 2, ctx=ctx)
self.block1 = Block(shared, ctx=ctx)
self.block2 = Block(shared, ctx=ctx)
def __call__(self, x, *, ctx: nnx.Context):
x = self.block1(x, ctx=ctx)
x = self.block2(x, ctx=ctx)
return x
In this example, the Model module contains two instances of the Block module. Each instance shares the same nnx.Linear module. To run the model, you can use the Context flags argument to set the use_running_average flag for all BatchNorm modules.
Here's an example of computing the loss for a Model instance:
def loss_fn(model: Model, x: jax.Array, y: jax.Array):
ctx = nnx.Context(flags=dict(use_running_average=True))
y_pred = model(x, ctx=ctx)
return jnp.mean((y - y_pred) ** 2)
It's important to note that the state for the shared nnx.Linear module will be kept in sync at all times on both Block instances, including during gradient updates.
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
Filter files by name, interpreter, ABI, and platform.
If you're not sure about the file name format, learn more about wheel file names.
Copy a direct link to the current filters
File details
Details for the file nnx-0.0.4.tar.gz.
File metadata
- Download URL: nnx-0.0.4.tar.gz
- Upload date:
- Size: 38.1 kB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: poetry/1.4.0 CPython/3.8.10 Linux/5.13.0-1027-gcp
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
4513efb1d586d503648035d43082c056f899974a876d87e65059b42e1be3f91f
|
|
| MD5 |
6682485d7e0e13118a0fb99adae5f42a
|
|
| BLAKE2b-256 |
0e2d0102a697abd4d128e6369fd8ac0d1f57aa07df32d351abd54ae0c9f04e8e
|
File details
Details for the file nnx-0.0.4-py3-none-any.whl.
File metadata
- Download URL: nnx-0.0.4-py3-none-any.whl
- Upload date:
- Size: 38.5 kB
- Tags: Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: poetry/1.4.0 CPython/3.8.10 Linux/5.13.0-1027-gcp
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
ea9818659c614f1014ea2ffa0485925f8342973a0c00b5892a8c55e3fab16ff3
|
|
| MD5 |
6a0a85cd852d1d24418706d570103f2c
|
|
| BLAKE2b-256 |
a9b658ff9e98aa724307870dc1f5e202d55822e80118a40445a93461ddd7467c
|