A Python plugin for OMERO.web
Project description


OMERO.gallery
This is an OMERO.web plugin (Django app) that provides a ‘gallery’ view of images in OMERO, ideal for public browsing without editing.
Also see SUPPORT.md
Requirements
OMERO.web 5.6.0 or newer.
Installing from PyPI
This section assumes that an OMERO.web is already installed.
Install the app using pip:
$ pip install -U omero-gallery
Add gallery custom app to your installed web apps:
$ omero config append omero.web.apps '"omero_gallery"'
Now restart OMERO.web as normal.
OMERO.gallery overview
This application supports 2 alternative views of your data in OMERO, which can be chosen and customised via config settings:
Default UI (no config): Browse Group > Project > Dataset > Image
Categories UI: Show categories of interest. Allow filtering by map annotations.
For both views, public access can be enabled as described here, otherwise users will see the standard web login screen. Once logged-in (as a regular user or public user), the data displayed will include all data accessible to that user via the normal OMERO permissions.
Default UI
This view supports minimal functionality required for browsing the hierarchy from Groups -> Projects -> Datasets -> Images. Screen/Plate/Well data is not supported in this UI.
The home page will display all the available groups that the user can access, with a random thumbnail from each group. The number of Projects, Datasets and Images within each group will also be displayed.
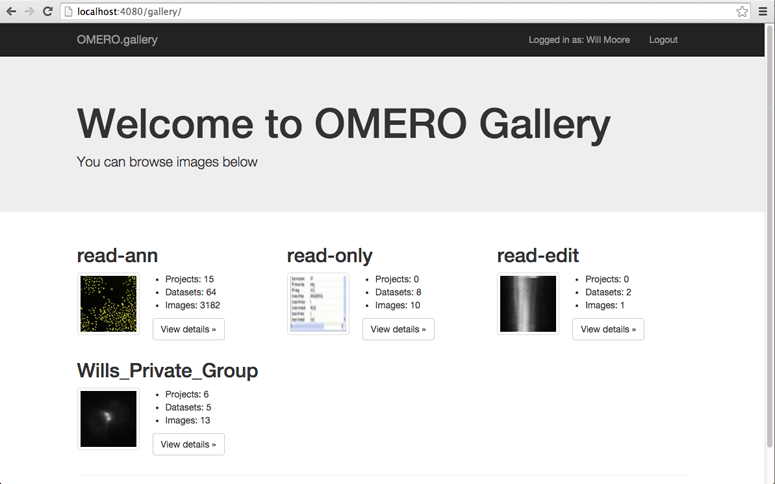
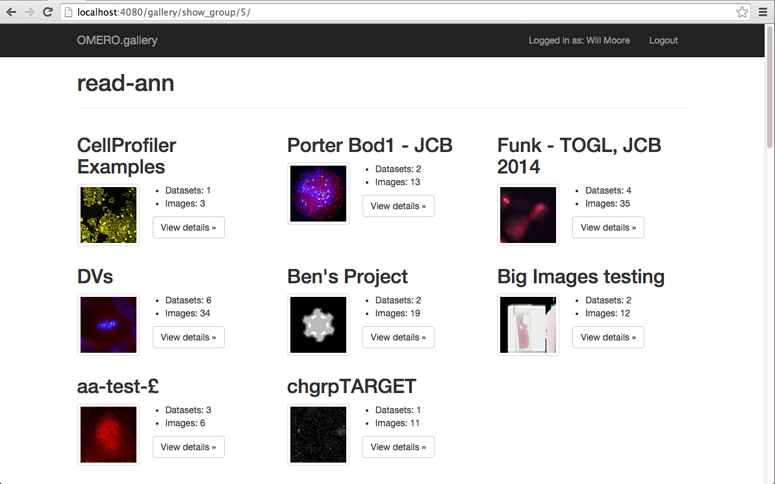
On browsing into a group, the Projects and ‘orphaned’ Datasets will be shown in a similar layout.
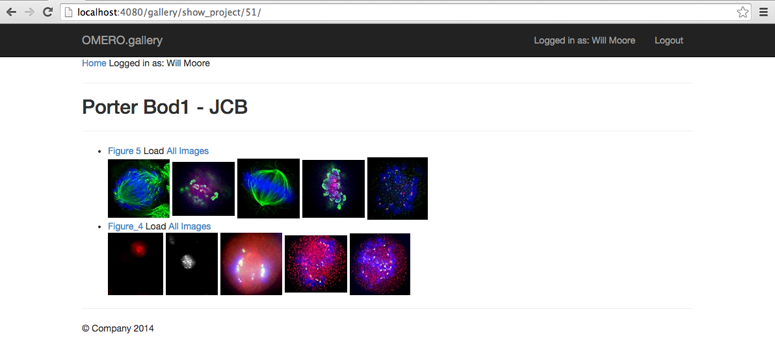
Projects are shown with 5 thumbnails from each Dataset. Clicking ‘All Images’ will load all the remaining thumbnails from a chosen Dataset (or you can browse to the Dataset itself by clicking the Dataset name link).
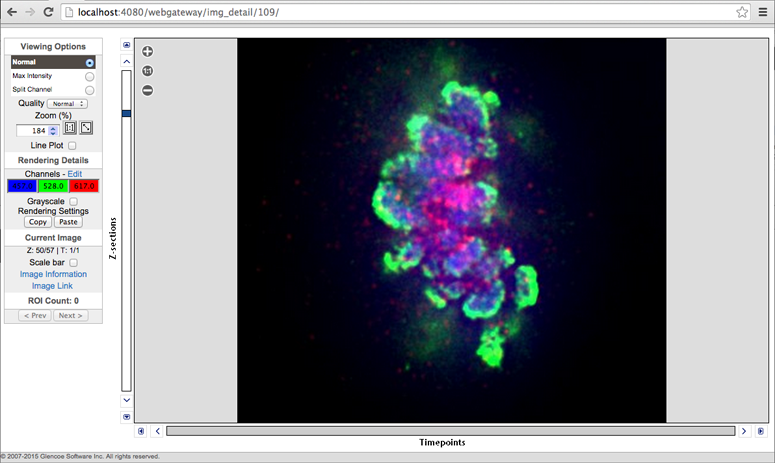
Clicking a thumbnail will take you directly to the full image viewer.
Categories UI
This view was originally developed for use in the IDR and can be seen at https://idr.openmicroscopy.org/. In the IDR, a “Study” is a Project or Screen and they are annotated with Key-Value data in the form of Map Annotations, for example Study Type: 3D-tracking. The UI supports several features based on these Key-Value attributes:
Home page shows ‘Categories’ that are defined by queries on Map Annotations.
Filter studies by Map Annotations.
If Images are also annotated with Map Annotations and https://github.com/ome/omero-mapr/ is installed then you can:
Find Studies containing Images that match queries on their Map Annotations.
Configuring the Categories UI
omero.web.gallery.category_queries: To enable the Categories UI, you must set omero.web.gallery.category_queries. If this is not set, you will see the Default UI shown above and the other settings described below will be ignored.
Each Category is defined by a display label, a query to select the Projects and Screens and an index to specify the order they appear on the page. Most of the examples below are used in the IDR. You can view the Categories at https://idr.openmicroscopy.org/ and see the query for each as a tooltip on the label of each category.
In the simplest case, if you do not have Map Annotations on Studies (Projects and Screens), you can simply sort by Name. This example defines a Category: “All Studies” to show the first 50 studies by Name:
$ omero config set omero.web.gallery.category_queries '{
"all":{"label":"All Studies", "index":0, "query":"FIRST50:Name"}
}'
Other categories are defined by queries on Map Annotations. For example, to show all Studies that have Key:Value of Study Type: 3D-tracking:
$ omero config set omero.web.gallery.category_queries '{
"tracking":{"label":"3D tracking", "index":0, "query":"Study Type: 3D-tracking"}
}'
Queries can use the AND and OR keywords to combine queries:
$ omero config set omero.web.gallery.category_queries '{
"time":{"label":"Time-lapse imaging", "index":0, "query":"Study Type: 3D-tracking OR Study Type: time"},
"screens":{"label":"High-content screening (human)", "index":1, "query":"Organism:Homo sapiens AND Study Type:high content screen"}
}'
omero.web.gallery.filter_keys: If this is configured then the gallery will allow filtering of Screens and Projects by Key:Value pairs linked to them, or use Name to filter by Name. This list defines which Keys the user can choose in the UI. On selecting a Key, the user will be able to filter by Values typed into an auto-complete field.
Each item is a simple string (matching the Key) or an object with a label and value, where value matches the Key. An example based on IDR:
$ omero config set omero.web.gallery.filter_keys '[
"Name",
"Imaging Method",
"Organism",
{"label": "Publication Authors", "value": "Authors"}
]'
omero.web.gallery.title: Sets the html page `<title>title</title>` for gallery pages.
omero.web.gallery.top_left_logo: This setting can be used to replace the ‘OMERO’ logo at the top-left of the page with an image hosted elsewhere (png, jpeg or svg). It will be displayed with height of 33 pixels and maximum width of 200 pixels:
$ omero config set omero.web.gallery.top_left_logo '{"src": "https://www.openmicroscopy.org/img/logos/ome-main-nav.svg"}'
omero.web.gallery.heading: Replace the “Welcome to OMERO.gallery” heading on the home page.
omero.web.gallery.top_right_links: This specifies a list of links as {‘text’:’Text’,’href’:’www.url’} for the top-right of each page. If a link contains ‘submenu’:[ ] with more links, these will be shown in a dropdown menu:
$ omero config set omero.web.gallery.top_right_links '[
{"text":"OME", "href":"https://www.openmicroscopy.org/"}
]'
omero.web.gallery.favicon: Set a URL to a favicon to use for the browser.
omero.web.gallery.subheading_html: Set some HTML to show as a sub-heading on the home page, within a <p> tag:
$ omero config set omero.web.gallery.subheading_html "This is an image gallery using <b>OMERO</b>."
omero.web.gallery.footer_html: Set some HTML to show as a footer on each page:
$ omero config set omero.web.gallery.footer_html "<a href='https://blog.openmicroscopy.org/'>Blog</a>"
omero.web.gallery.study_short_name: This specifies a short name for Screen or Project to show above the study Image in the categories or search page, instead of the default ‘Project: 123’. The list allows us to try multiple methods, using the first that works. Each object in the list has e.g. {‘key’: ‘Name’}. The ‘key’ can be Name, Description or the key for a Key:Value pair on the object. If a ‘regex’ and ‘template’ are specified, we try name.replace(regex, template). In this example, we check for a Key:Value named “Title”. If that is not found, then we use a regex based on the object’s Name. This example is from the IDR, where we want to create a short name like idr0001A from a Name like: idr0001-graml-sysgro/screenA:
$ omero config set omero.web.gallery.study_short_name '[
{"key":"Title"},
{"key":"Name", "regex": "^(.*?)-.*?(.)$", "template": "$1$2"},
]'
License
OMERO.gallery is released under the AGPL.
Copyright
2016-2020, The Open Microscopy Environment
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
File details
Details for the file omero-gallery-3.3.0.tar.gz.
File metadata
- Download URL: omero-gallery-3.3.0.tar.gz
- Upload date:
- Size: 298.9 kB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/3.1.1 pkginfo/1.5.0.1 requests/2.22.0 setuptools/45.0.0 requests-toolbelt/0.9.1 tqdm/4.41.1 CPython/3.6.7
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | a6c6e640286580d02c91944776d8d105a96653d17f0c769fbbe8cb93a1c36205 |
|
| MD5 | 3c2c788b19528e47f705a19d0eaad578 |
|
| BLAKE2b-256 | 58405d84dcd2f960ae2022ee73ef76879f1a8bc92d7494529896324eafe4d9f1 |
File details
Details for the file omero_gallery-3.3.0-py3-none-any.whl.
File metadata
- Download URL: omero_gallery-3.3.0-py3-none-any.whl
- Upload date:
- Size: 307.4 kB
- Tags: Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/3.1.1 pkginfo/1.5.0.1 requests/2.22.0 setuptools/45.0.0 requests-toolbelt/0.9.1 tqdm/4.41.1 CPython/3.6.7
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 5cb48079e5e3c39818fcab8a4444221f9fd6b8c0c3771a4e67bed113385e0059 |
|
| MD5 | 0a754bed9b4f32541bdb3f5864738960 |
|
| BLAKE2b-256 | 0417ce87d272080f0f795d01bb7c48f091fc2e92911ddcb33841382db8e5353c |














