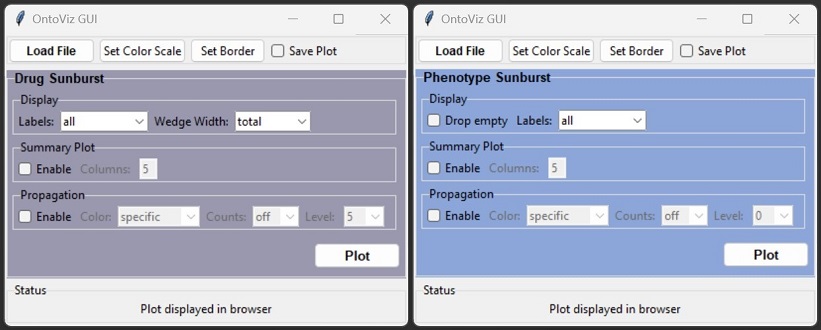
OntoloViz drug- and phenotype-ontology visualization GUI
Project description
OntoloViz
OntoloViz is a graphical user interface for the creation of interactive sunburst plots of phenotype and drug ontologies.
You might find it useful to quickly visualize your data for reports or to share the generated plots with collaborators.
Take a look at the screenshot section or explore examples in .html format from the
provided templates.zip archive to get a
better understanding of the scope of the package.
Quickstart
The GUI can be run by downloading the latest Windows release or by installing and running the package via PyPi (OS independent, requires Python 3.10+):
pip install ontoloviz
python -c "from ontoloviz import run_app; run_app()"
Alternatively, you can clone this repository, install the required dependencies and launch the GUI:
git clone https://github.com/Mnikley/OntoloViz.git
cd OntoloViz
pip install -r requirements.txt
python src/ontoloviz/app.py
Usage
The application allows importing .tsv and .xlsx files, but the use of .tsv and tab as a
separator is recommended. The GUI can create two types of sunburst diagrams to represent either phenotype or drug
ontologies, which is determined by the structure of the loaded files. Any numbers entered in the file will be converted
to integers.
GUI Options
- General
- Load File: load an
.tsvor.xlsxfile containing drug- or phenotype-ontology data - Set Color Scale: define a custom color scale for the sunburst color scaling when color propagation is active
- Set Border: configures the border properties drawn around sunburst wedges
- Save Plot: when enabled, an interactive
.htmlfile is generated for later use
- Load File: load an
- Display
- Drop empty (phenotype sunburst only): drops nodes who have no further children and 0 counts
- Labels: controls display of labels inside sunburst wedges, available options:
allpropagationdrugs(drug sunburst only)none
- Summary Plot
- Enable: displays all available subtrees in a single view (resource intensive, set Labels to
nonefor faster loading) - Columns: defines the amount of columns when summary plot is enabled
- Enable: displays all available subtrees in a single view (resource intensive, set Labels to
- Propagation
- Enable: enables count- and color propagation from child to parent nodes
- Color: controls color propagation by the options:
off: color scale is based on 'Color' column from imported filespecific: color scale is based on the maximum values of the corresponding subtreeglobal: color scale is based on the maximum values of the entire tree ontologyphenotype(phenotype sunburst only): Only the most outer phenotype in a branch is colored
- Counts: controls count propagation by the options:
off: no counts are propagated, counts equal imported valueslevel: counts are propagated up to defined level, values above threshold remain unchangedall: counts are propagated up to central node, imported values are corrected and overwritten
- Level: controls color- and count-propagation from outer to inner levels up to defined level
- affects color propagation when Color is set to
specificorglobal - affects count propagation when Counts is set to
level - drug sunburst: 1 corresponds to the central node, 5 to the outermost node (=drug)
- phenotype sunburst: 0 corresponds to the central node, 13 to the outermost node
- affects color propagation when Color is set to
Phenotype Sunbursts
The phenotype sunburst structure follows the principles of the MeSH tree.
- A Tree ID is defined by a combination of three numbers or letters, for example
C01. - Levels are separated by a dot
., for exampleC01.001. - Ontologies up to thirteen hierarchical levels are supported.
- A single phenotype end-node can be assigned to multiple parent-nodes by specifying the parents tree ids as
pipe separated string in the column
Tree ID. - When defining a child element which has no valid parent, the GUI will automatically generate the parent with the
default color and a 0 value. This will happen recursively. For example, if the input file defines a node with the
id
123.001.001, but the nodes123and123.001are non-existent, they will be created. - Counts entered in the file will be converted to integers. If a node should be displayed without counts, use
0. - The loaded file must contain 7 columns and follow the below structure to be correctly recognized:
Phenotype Ontology File Structure
| Column Index | Header Text | Description |
|---|---|---|
| 0 | MeSH ID | Required primary identifier of a node in format C01.001 |
| 1 | Tree ID | Required pipe delimited list of Tree IDs of a node (allows 1:N mappings) |
| 2 | Name | Optional label to be displayed inside the sunburst wedges |
| 3 | Description | Optional description displayed in the sunburst wedge tooltip |
| 4 | Comment | Optional comment displayed in the sunburst wedge tooltip |
| 5 | Counts [Name] | Required count for wedge weights, Name will be used as figure title |
| 6 | Color | Optional color for the sunburst wedges, must be hex-string in format #FFFFFF |
Drug Sunbursts
The drug sunburst structure follows the principles of the ATC tree.
- ATC codes are divided into five levels, which must follow the following naming conventions:
- 1st level: letter
- 2nd level: two numbers
- 3rd level: letter
- 4th level: letter
- 5th level: two numbers
- Example ATC code: A10BA02
- The hierarchy is built based on the above-mentioned format and does only allow 1:1 child-parent relationships
(contrary to the phenotype structure). For example, if the drug
deltatoninshould be assigned to the parent nodesA01AAandB01BB, it must be defined twice with the idsA01AA01andB01BB01. - The loaded file must contain 6 columns and follow the below structure to be correctly recognized as a phenotype ontology:
Drug Ontology File Structure
| Column Index | Header Text | Description |
|---|---|---|
| 0 | ATC code | Required primary identifier of a node in format A10BA02 |
| 1 | Level | Optional level as number, not used for building tree |
| 2 | Label | Optional label to be displayed inside the sunburst wedges |
| 3 | Comment | Optional comment displayed in the sunburst wedge tooltip |
| 4 | Counts [Name] | Required count for wedge weights, Name will be used as figure title |
| 5 | Color | Optional color for the sunburst wedges, must be hex-string in format #FFFFFF |
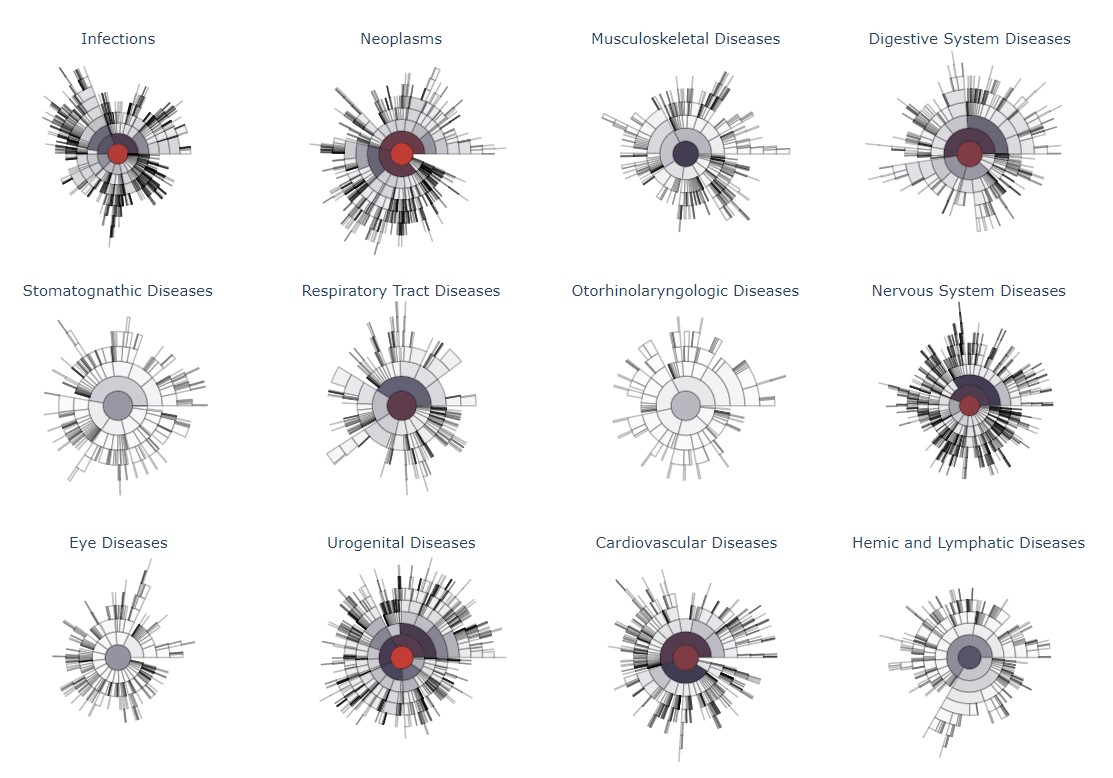
Screenshots
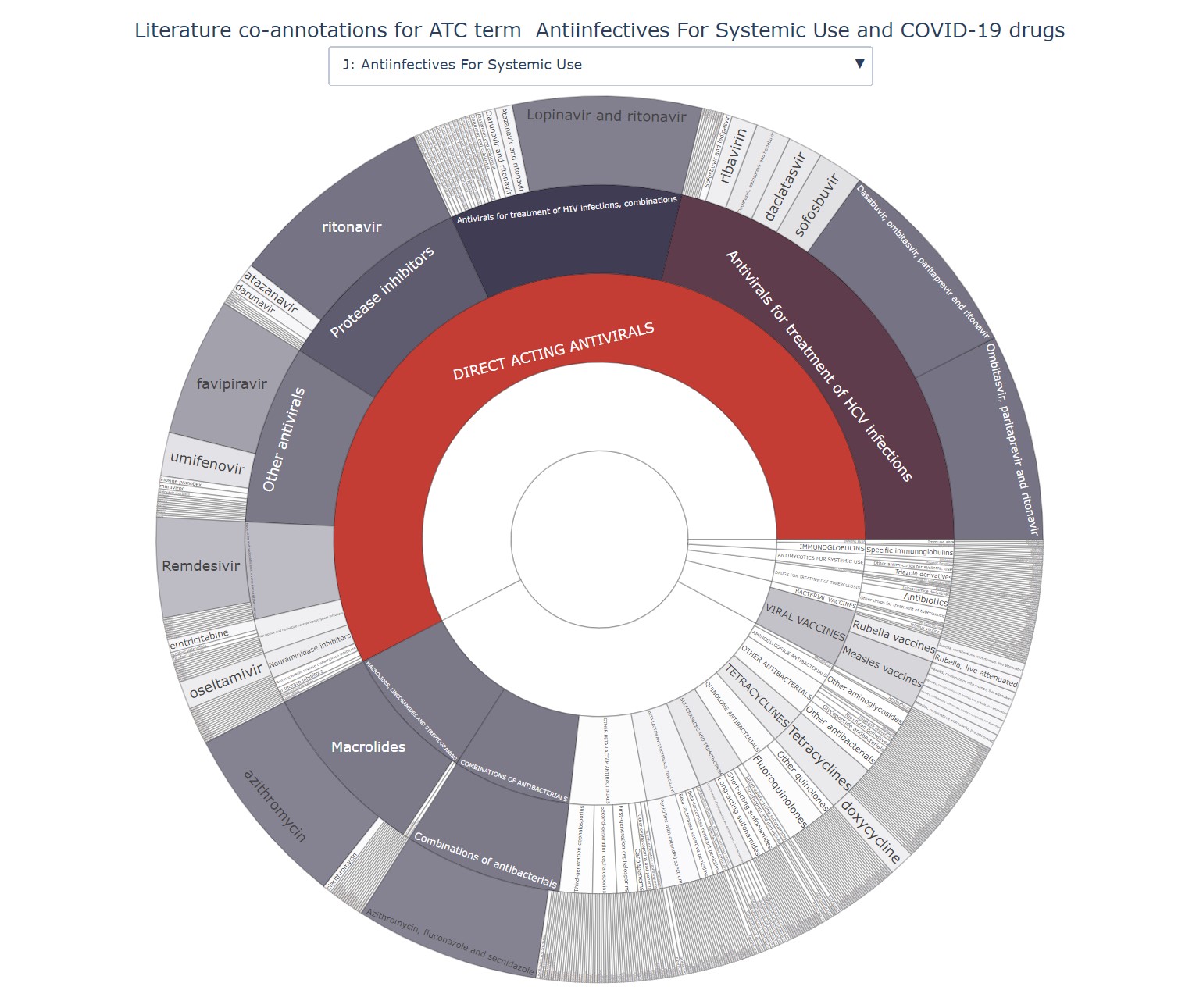
Templates and Examples
Templates and examples can be found in the provided templates.zip archive.
-
pubmed_documents_mapped_to_mesh.tsv: based on the MeSH subtreeCfrom 2022. Disease-related MeSH terms were extracted from the publicly available PubMed database (title + abstract) and further mapped to the nodes. -
mesh_tree_template.tsv: empty template of the MeSH treeCandF03]. Terms are unique and mapped to all related parent nodes. -
covid_drugs_trial_summary.tsv: based on publicly available clinical trial data related to COVID-19. One count represents one clinical trial. -
atc_tree_template.tsv: empty template of the ATC tree based on the manually curated chemical database of bioactive molecules ChEMBL v29. -
drug_sunburst_example.html: sample plot generated with the providedcovid_drugs_trial_summary.tsvfile. -
phenotype_sunburst_example.html: sample plot generated with the providedcovid_drugs_trial_summary.tsvfile.
Special Thanks to
- Paul Perco, who had the initial idea for this package and provided support throughout the process
- Andreas Heinzel, who is an overall inspiration regarding all software- and non-software related topics
- The Delta4 GmbH team for providing various helpful inputs
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
File details
Details for the file ontoloviz-1.3.3.tar.gz.
File metadata
- Download URL: ontoloviz-1.3.3.tar.gz
- Upload date:
- Size: 40.6 kB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/4.0.2 CPython/3.11.0
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 1404381f542b85c3a5d92b831556967975d280f238134254583acfa17e8b0f0b |
|
| MD5 | 7f3454f1c7fa0c40832613f4e7db8ca9 |
|
| BLAKE2b-256 | 756d482536cd5cb7540a93edc324e0c6479b621e81232241214d1d1792754335 |
File details
Details for the file ontoloviz-1.3.3-py3-none-any.whl.
File metadata
- Download URL: ontoloviz-1.3.3-py3-none-any.whl
- Upload date:
- Size: 37.7 kB
- Tags: Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/4.0.2 CPython/3.11.0
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | c1e38e64afba69d1af8f2ec2f7e96ef41f2db203c007df115e219d3bdd5d72b0 |
|
| MD5 | 6f8a1ad69fc24fe66fc5504a3ab080d0 |
|
| BLAKE2b-256 | 6306e6d44e73782d704f3c72672a1c026f006ad307ad7e2c8431d1d771c48abd |