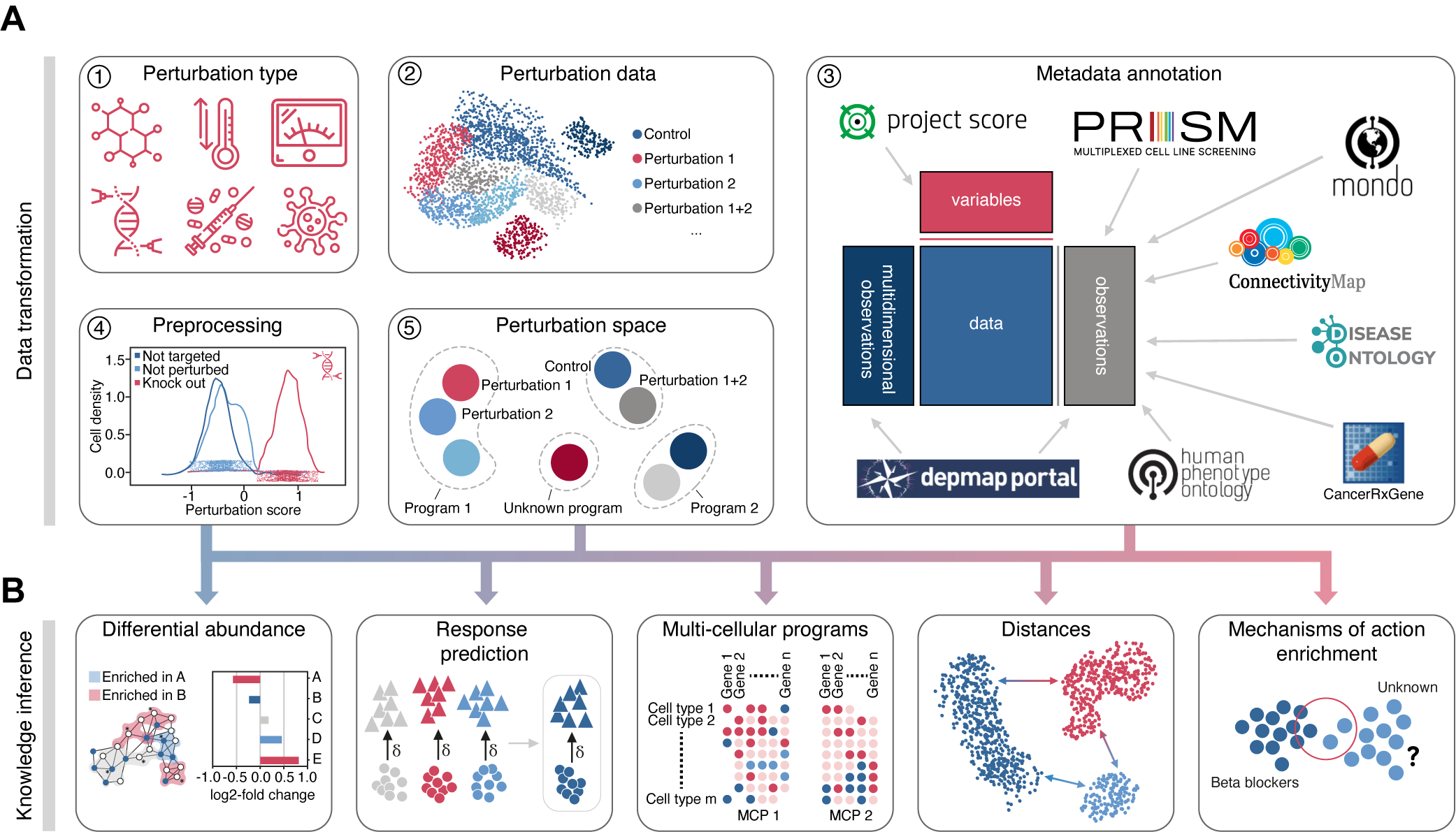
Perturbation Analysis in the scverse ecosystem.
Project description
pertpy - Perturbation Analysis in Python
Pertpy is a scverse ecosystem framework for analyzing large-scale single-cell perturbation experiments. It provides tools for harmonizing perturbation datasets, automating metadata annotation, calculating perturbation distances, and efficiently analyzing how cells respond to various stimuli like genetic modifications, drug treatments, and environmental changes.
Documentation
Please read the documentation for installation, tutorials, use cases, and more.
Installation
We recommend installing and running pertpy on a recent version of Linux (e.g. Ubuntu 24.04 LTS). No particular hardware beyond a standard laptop is required.
You can install pertpy in less than a minute via pip from PyPI:
pip install pertpy
or conda-forge:
conda install -c conda-forge pertpy
Differential gene expression
If you want to use the differential gene expression interface, please install pertpy by running:
pip install 'pertpy[de]'
tascCODA
if you want to use tascCODA, please install pertpy as follows:
pip install 'pertpy[tcoda]'
milo
milo requires either the "de" extra for the "pydeseq2" solver:
pip install 'pertpy[de]'
or, edger, statmod, and rpy2 for the "edger" solver:
BiocManager::install("edgeR")
BiocManager::install("statmod")
pip install rpy2
Citation
@article{Heumos2025,
author = {Heumos, Lukas and Ji, Yuge and May, Lilly and Green, Tessa D. and Peidli, Stefan and Zhang, Xinyue and Wu, Xichen and Ostner, Johannes and Schumacher, Antonia and Hrovatin, Karin and Müller, Michaela and Chong, Faye and Sturm, Gregor and Tejada, Alejandro and Dann, Emma and Dong, Mingze and Pinto, Gonçalo and Bahrami, Mojtaba and Gold, Ilan and Rybakov, Sergei and Namsaraeva, Altana and Moinfar, Amir Ali and Zheng, Zihe and Roellin, Eljas and Mekki, Isra and Sander, Chris and Lotfollahi, Mohammad and Schiller, Herbert B. and Theis, Fabian J.},
title = {Pertpy: an end-to-end framework for perturbation analysis},
journal = {Nature Methods},
year = {2025},
publisher = {Springer Nature},
doi = {10.1038/s41592-025-02909-7},
url = {https://doi.org/10.1038/s41592-025-02909-7},
issn = {1548-7105}
}
pertpy is part of the scverse® project (website, governance) and is fiscally sponsored by NumFOCUS. If you like scverse® and want to support our mission, please consider making a tax-deductible donation to help the project pay for developer time, professional services, travel, workshops, and a variety of other needs.
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
Filter files by name, interpreter, ABI, and platform.
If you're not sure about the file name format, learn more about wheel file names.
Copy a direct link to the current filters
File details
Details for the file pertpy-1.0.5.tar.gz.
File metadata
- Download URL: pertpy-1.0.5.tar.gz
- Upload date:
- Size: 4.1 MB
- Tags: Source
- Uploaded using Trusted Publishing? Yes
- Uploaded via: twine/6.1.0 CPython/3.13.7
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
9f78eaf1ee6231b7c2e9820a2a9247d4dabc1d04674062f1f5ca2ffe382fb310
|
|
| MD5 |
76de2e54a75bf5f9d97aa33d50112fa4
|
|
| BLAKE2b-256 |
80eba0b1b9ff17351bd769efc6a3d412ee2fd35d99c4b246724e52b3d2d18c41
|
Provenance
The following attestation bundles were made for pertpy-1.0.5.tar.gz:
Publisher:
release.yml on scverse/pertpy
-
Statement:
-
Statement type:
https://in-toto.io/Statement/v1 -
Predicate type:
https://docs.pypi.org/attestations/publish/v1 -
Subject name:
pertpy-1.0.5.tar.gz -
Subject digest:
9f78eaf1ee6231b7c2e9820a2a9247d4dabc1d04674062f1f5ca2ffe382fb310 - Sigstore transparency entry: 862821376
- Sigstore integration time:
-
Permalink:
scverse/pertpy@efe9d552b4d46a7ed3cf8913c96ea58623f384f0 -
Branch / Tag:
refs/tags/1.0.5 - Owner: https://github.com/scverse
-
Access:
public
-
Token Issuer:
https://token.actions.githubusercontent.com -
Runner Environment:
github-hosted -
Publication workflow:
release.yml@efe9d552b4d46a7ed3cf8913c96ea58623f384f0 -
Trigger Event:
release
-
Statement type:
File details
Details for the file pertpy-1.0.5-py3-none-any.whl.
File metadata
- Download URL: pertpy-1.0.5-py3-none-any.whl
- Upload date:
- Size: 225.2 kB
- Tags: Python 3
- Uploaded using Trusted Publishing? Yes
- Uploaded via: twine/6.1.0 CPython/3.13.7
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
f305decce41460ed20d13dffd7df6ff1c56596db850989d87c5a2326641db9a9
|
|
| MD5 |
685f5212fdbe28343be181f96339d946
|
|
| BLAKE2b-256 |
a47ba0682f9e08a58a24bb4cd3a2272d348f6e8d27b36f66477dc4c3c0a544c5
|
Provenance
The following attestation bundles were made for pertpy-1.0.5-py3-none-any.whl:
Publisher:
release.yml on scverse/pertpy
-
Statement:
-
Statement type:
https://in-toto.io/Statement/v1 -
Predicate type:
https://docs.pypi.org/attestations/publish/v1 -
Subject name:
pertpy-1.0.5-py3-none-any.whl -
Subject digest:
f305decce41460ed20d13dffd7df6ff1c56596db850989d87c5a2326641db9a9 - Sigstore transparency entry: 862821403
- Sigstore integration time:
-
Permalink:
scverse/pertpy@efe9d552b4d46a7ed3cf8913c96ea58623f384f0 -
Branch / Tag:
refs/tags/1.0.5 - Owner: https://github.com/scverse
-
Access:
public
-
Token Issuer:
https://token.actions.githubusercontent.com -
Runner Environment:
github-hosted -
Publication workflow:
release.yml@efe9d552b4d46a7ed3cf8913c96ea58623f384f0 -
Trigger Event:
release
-
Statement type: