QIIME 2 plugin for metabolic modeling of microbial communities.
Project description
A QIIME 2 plugin for MICOM.
Installation
You will need an existing QIIME 2 environment. Follow the instructions on (how to install QIIME 2)
otherwise. Let's assume that environment was called qiime2-2021.2 for all further steps.
Add q2-micom to the QIIME 2 environment
This will be the same step for any supported QIIME 2 version or operating systems.
wget https://raw.githubusercontent.com/micom-dev/q2-micom/master/q2-micom.yml
conda env update -n qiime2-2021.2 -f q2-micom.yml
# OPTIONAL CLEANUP
rm q2-micom.yml
Finally, you activate your environment.
conda activate qiime2-2021.2
Install a QP solver
CPLEX (recommended)
QIIME 2 versions before 2021.4 are only compatible with CPLEX 12.10 or earlier (later version require at least Python 3.7).
After registering and downloading the CPLEX studio for your OS unpack it (by running the provided installer) to a directory of your choice (we will assume it's called ibm).
Now install the CPLEX python package into your activated environment:
pip install ibm/cplex/python/3.6/x86-64_linux
Substitute 3.6 with the Python version in your QIMME 2 environment, 3.6 for QIIME 2 up to 2021.2 and 3.8 for QIIME 2 2021.4 and newer.
Substitute x86-64_linux with the folder corresponding to your system (there will only be one subfolder in that directory).
Gurobi (works, but not recommended)
q2-micom is not tested against Gurobi. Consequently Gurobi support is often iffy and might break for periods of time. It will also be much slower than CPLEX or OSQP.
You should only consider using Gurobi if:
- You do not have access to CPLEX
- You do need high accuracy solutions (tolerance of 1e-6 or lower)
Gurobi can be installed with conda.
conda install -c gurobi gurobi
You will now have to register the installation using your license key.
grbgetkey YOUR-LICENSE-KEY
Finish your installation
If you installed q2-micom in an already existing QIIME 2 environment, update the plugin cache:
conda activate qiime2-2021.2 # or whatever you called your environment
qiime dev refresh-cache
You are now ready to run q2-micom!
Usage
Here is a graphical overview of a q2-micom analysis.
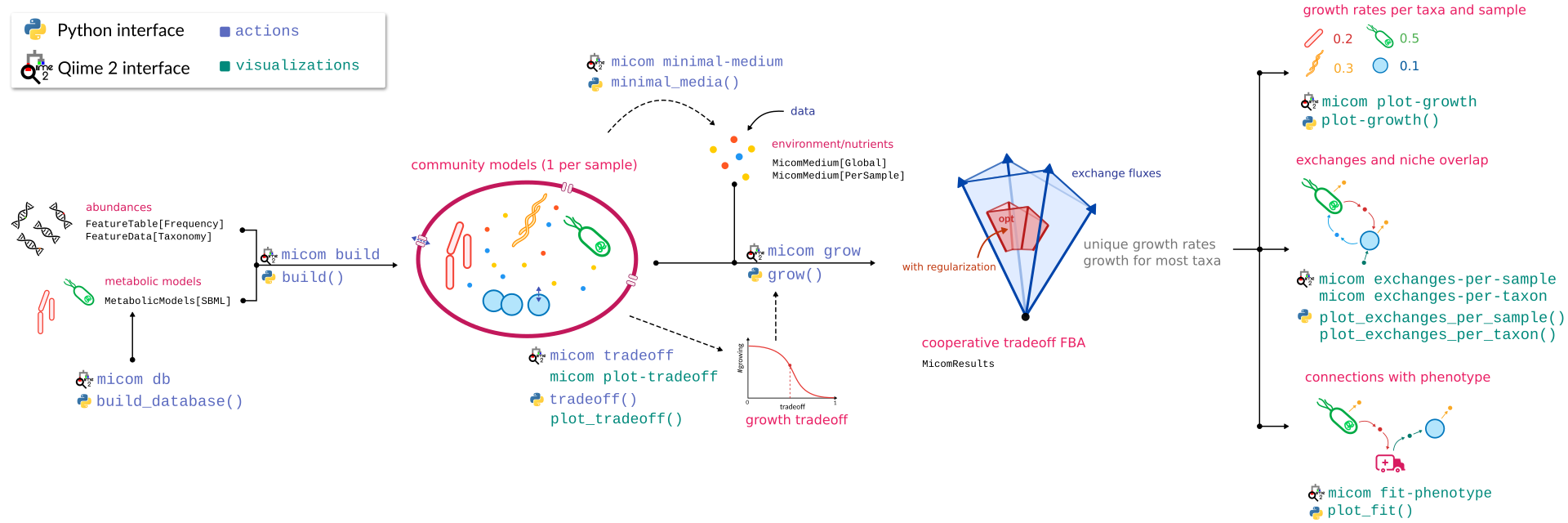
The best way to get started is to work through the community tutorial.
Supported QIIME 2 versions
q2-micom is tested against:
- the current QIIME 2 version
- the previous version
It should also work with
- the development version
However, this may occasionally break. Check here for the current status.
References
MICOM: Metagenome-Scale Modeling To Infer Metabolic Interactions in the Gut Microbiota
Christian Diener, Sean M. Gibbons, Osbaldo Resendis-Antonio
mSystems 5:e00606-19
https://doi.org/10.1128/mSystems.00606-19
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
Hashes for q2_micom-0.11.3-py3-none-any.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | fb0831170df5204405eaae9a7bb426ef4f98c72b9c8539a0acd26defc2c88d01 |
|
| MD5 | cdc38199188c640233ccf13b266bb030 |
|
| BLAKE2b-256 | 9def8cc25e10e19d67944298afc63ac3ac989098405eb888e92c9790a0fbb07b |














