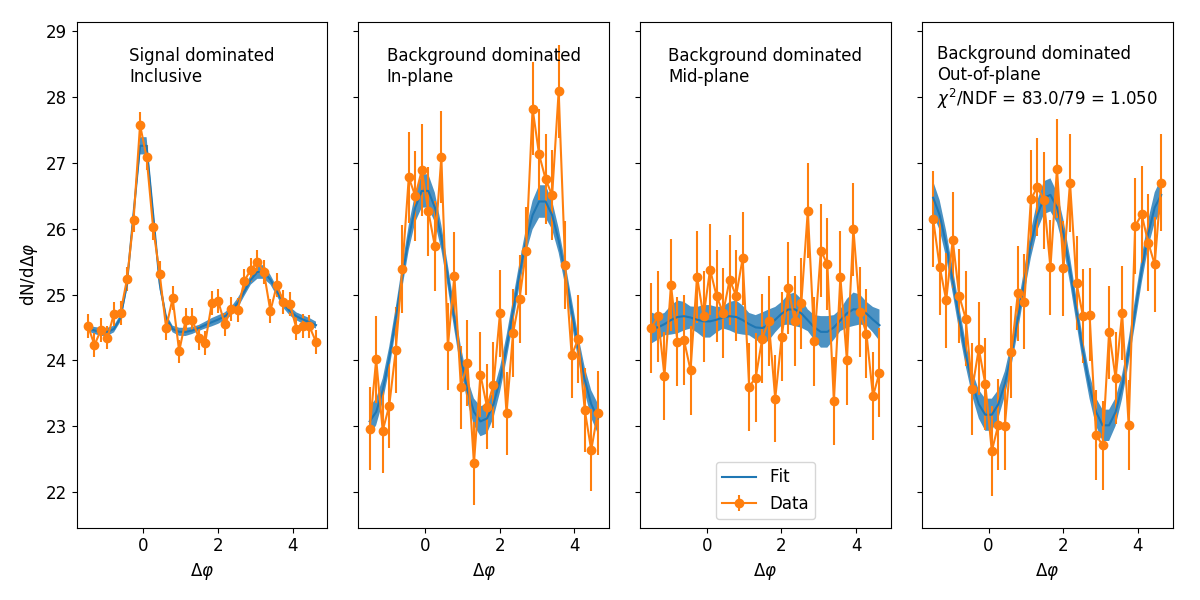
Implments the reaction plane fit for background subtraction in heavy ion collisions.
Project description
Reaction plane fit
Implements the reaction plane (RP) fit described in Phys. Rev. C 93, 044915 (or on the arxiv) to characterize and subtract background contributions to correlation functions measured in heavy ion collisions. It allows for fits to the background dominated region, as well as the option of fitting the RP inclusive signal dominated region or the RP dependent signal dominated region. This package implements the fit for three orientations relative to the RP, with the possibility of straightforward extension to other sets of orientations.
Installation
This package requires python 3.6 and above. A few prerequisites are required which unfortunately cannot be
resolved solely by pip because of the packaging details of probfit.
$ pip install numpy cython
The package is available on PyPI and is available via pip.
$ pip install reaction_plane_fit
You may want to consider installing this to your user directory (--user) or better, within a virtual
environment.
Usage
Performing a fit with this package only requires a few lines of code. Below is sufficient to define and run a fit with some sample values:
from reaction_plane_fit import three_orientations
# Define the fit object.
rp_fit = three_orientations.BackgroundFit(
resolution_parameters = {"R22": 0.6, "R42": 0.3, "R62": 0.1, "R82": 0.1},
use_log_likelihood = False,
signal_region = (0, 0.6),
background_region = (0.8, 1.2),
)
# Load or otherwise provide the relevant histograms here.
# The structure of this dictionary is important to ensure that the proper data ends up in the right place.
# Note that the data must already be projected into the background or signal dominated regions.
data = {"background": {"in_plane": ROOT.TH1.., "mid_plane": ROOT.TH1..., "out_of_plane": ROOT.TH1...}}
# Perform the actual fit.
success, _ = rp_fit.fit(data = data)
# Print the fit results
print("Fit result: {fit_result}".format(fit_result = rp_fit.fit_result))
Examples for fitting an inclusive reaction plane orientation signal alongside the background, or for fitting
only the background are both available in reaction_plane_fit.example. This module can also be run directly
in the terminal via:
$ python -m reaction_plane_fit.example [-b] [-i dataFilename]
If fit data is not specified, it will use some sample data. For further information, including all possible fit function combinations, please see the full documentation.
But I don't like python!
You might not be so happy about using Python. That's okay - we can work around this using ROOT, although it will be much more painful than using python directly. To do so, it should look something like (the code below is untested, so it may need some minor modifications):
// Setup the input data.
std::map<std::string, std::map<std::string, TH1*>> inputData;
inputData["background"]["in_plane"] = my_in_plane_hist;
// Fill in the rest of your input data.
...
// Setup the fit objects.
TPython::Exec("from reaction_plane_fit import three_orientations");
TPython::Exec("rp_fit = three_orientations.BackgroundFit("
"resolution_parameters = {'R22': 0.6, 'R42': 0.3, 'R62': 0.1, 'R82': 0.1},"
"use_log_likelihood = False,"
"signal_region = (0, 0.6),"
"background_region = (0.8, 1.2),)"
);
// Perform the actual fit.
TPython::Exec("success, _ = rp_fit.fit(data = data)");
// Print the fit results.
TPython::Exec("print('Fit result: {fit_result}'.format(fit_result = rp_fit.fit_result))")
// Access values back in c++.
int chi_2 = double(TPython::Eval("rp_fit.fit_result.minimum_val"))
// It will require some attention to extract all of the relevant values.
// You can extract a number of types (see TPyResult), but it doens't appear that you can extract complex objects.
// So this could still be a somewhat painful process.
(Don't forgot that we're calling Minuit underneath, so this package should be fast enough for this fit).
Fits implemented
There are three possible types of fits:
- Background dominated region only: This fits only regions on the near-side at large dEta which are dominated
by background contributions. Called
BackgroundFitin each set of orientations implementation. - Inclusive signal region: This fits to the RP inclusive signal dominated region, as well as fitting the RP
dependent background dominated regions. Called
InclusiveSignalFitin each set of orientations implementation. - RP dependent signal region: This fits to the RP dependent signal dominated region, as well as fitting the RP
dependent background dominated regions. Called
SignalFitin each set of orientations implementation.
Three orientation (in-, mid-, and out-of-plane)
This package implements the fit for three orientations relative to the reaction plane:
- in-plane (0<|Δφ|<π/6)
- mid-plane (π/6<|Δφ|<π/3)
- and out-of-plane (π/3<|Δφ|<π/2)
These fits are implemented in the reaction_plane_fit.three_orientations module.
Notes on fitting
This package makes it possible to use Minos rather than Hesse errors. Minos errors can be calculated when Hesse errors become inaccurate (when the function around the minima is not approximately a hyperparabola), but they can take much longer to calculate and cannot be described via a covariance matrix (which makes error propagation much more difficult). If the Hesse and Minos errors are similar, then the function is well approximately by a hyperparabola and you can safely use Hesse errors. For more, see the iminuit tutorials.
Development
If developing the packaging, clone the repository and then install with
$ pip install -e .[dev,tests]
Note that python 3.6 and above is required because this package uses dataclasses (which has a python 3.6
backport), and it relies on dictionaries being ordered (which is true for cpython 3.6 and is required for
python 3.7 in general).
Citation
Please cite the paper (Phys. Rev. C 93, 044915), as well as this implementation:
@misc{raymond_ehlers_2018_1599239,
author = {Raymond Ehlers},
title = {reactionPlaneFit - RPF implementation},
month = nov,
year = 2018,
doi = {10.5281/zenodo.1599239},
url = {https://doi.org/10.5281/zenodo.1599239}
}
Acknowledgments
Code started from implementation work done by M. Arratia. Thanks to C. Nattrass and J. Mazer for help and discussions.
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
File details
Details for the file reaction_plane_fit-2.0.tar.gz.
File metadata
- Download URL: reaction_plane_fit-2.0.tar.gz
- Upload date:
- Size: 39.7 kB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/1.13.0 pkginfo/1.5.0.1 requests/2.21.0 setuptools/40.8.0 requests-toolbelt/0.9.1 tqdm/4.31.1 CPython/3.7.1
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 8aa3c1ee9f3a2a5b801ea1e66fbf27c7040c93ec03544904a876f49d9cd8d62d |
|
| MD5 | 31bc0d462f3b8bdb7564f06bc9fccadb |
|
| BLAKE2b-256 | 8a2c06fd1cfe5db114130590008e0a439d7da5552aaa24ad9f0d29fa994e1568 |
File details
Details for the file reaction_plane_fit-2.0-py2.py3-none-any.whl.
File metadata
- Download URL: reaction_plane_fit-2.0-py2.py3-none-any.whl
- Upload date:
- Size: 39.2 kB
- Tags: Python 2, Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/1.13.0 pkginfo/1.5.0.1 requests/2.21.0 setuptools/40.8.0 requests-toolbelt/0.9.1 tqdm/4.31.1 CPython/3.7.1
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 21dcb173cb1683027f4889ad1a11946a218c8647a7b19473451b3214f61ac35d |
|
| MD5 | 47ac4f814469b124184ef52e0a28d5de |
|
| BLAKE2b-256 | 37fdd54b01c97ef00bd36c829768fa2c05916354784f22d1a81b889c79582274 |