Provides DNA overhang misannealing data (Potapov 2018).
Project description
Tatapov


Tatapov is a Python library making accessible and easy to explore the DNA overhang misannealing data from the following paper (available on arxiv):
Optimization of Golden Gate assembly through application of ligation sequence-dependent fidelity and bias profiling, Vladimir Potapov, Jennifer L. Ong, Rebecca B. Kucera, Bradley W. Langhorst, Katharina Bilotti, John M. Pryor, Eric J. Cantor, Barry Canton, Thomas F. Knight, Thomas C. Evans Jr., Gregory Lohman. May 2018, https://doi.org/10.1101/322297
The Supplementary Material of this paper provides tables of inter-overhang annealing data in various 4 conditions (01h or 18h incubations at 25C or 37C). Tatapov provides these tables (it will download them automatically from Arxiv on the first use) as Pandas dataframes, so that they are easy to manipulate.
It also provides simple methods to build and plot subsets of the data (plotting requires Matplotlib installed).
Usage Example
Plotting
import tatapov
# Get a subset of the data at 25C (1h incubation)
data = tatapov.annealing_data["25C"]["01h"] # a pandas dataframe
overhangs = ["ACGA", "AAAT", "AGAG"]
subset = tatapov.data_subset(data, overhangs, add_reverse=True)
# Plot the data subset
ax, _ = tatapov.plot_data(subset, figwidth=5)
ax.figure.tight_layout()
ax.figure.savefig("example.png")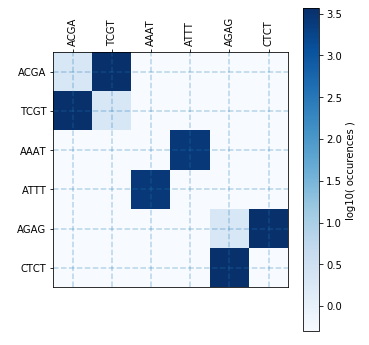
In the plot above, if you see anything else than the square pairs around the diagonal, it means there is cross-talking between your overhangs (so risk of misannealing). If one of these diagonal square pairs appears lighter than the others, it means that the corresponding overhang has weak self-annealing (risk of having no assembly). A color square in the diagonal means that the overhang can anneal with itself (palindromic).
Identifying weak self-annealing overhangs
import tatapov
annealing_data = tatapov.annealing_data['37C']['01h']
# Compute a dictionary {overhang: self-annealing score in 0-1}
relative_self_annealing = tatapov.relative_self_annealings(annealing_data)
weak_self_annealing_overhangs = [
overhang
for overhang, self_annealing in relative_self_annealing.items()
if self_annealing < 0.4
]Identifying overhang pairs with significant cross-talking
import tatapov
annealing_data = tatapov.annealing_data['37C']['01h']
# Compute a dictionary {overhang_pair: cross-talking score in 0-1}
cross_annealings = tatapov.cross_annealings(annealing_data)
high_cross_annealing_pairs = [
overhang_pair
for overhang_pair, cross_annealing in cross_annealings.items()
if cross_annealing > 0.08
]Installation
You can install Tatapov through PIP:
sudo pip install tatapovAlternatively, you can unzip the sources in a folder and type
sudo python setup.py installLicense = MIT
Tatapov is an open-source software originally written at the Edinburgh Genome Foundry by Zulko and released on Github under the MIT licence (Copyright 2018 Edinburgh Genome Foundry). Everyone is welcome to contribute !
Please contact us if there is any issue regarding copyright (there shouldn’t be as the repository does not contain any data, and the paper data is free to download).
More biology software

Tatapov is part of the EGF Codons synthetic biology software suite for DNA design, manufacturing and validation.
Project details
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.











