No project description provided
Project description
Tumor/T cell Microenvironment ABM
This project provides a multiscale agent-based model of the cancer microenvironment based on decades of research, lab-derived values, and using the backbone of Vivarium which is a flexible python-based framework that is extendable. The model is based on individual agents (e.g., cancer cells, immune cells) and their intracellular and intercellular interactions with both membrane bound and soluble factors (e.g., cytokines, cytotoxic granules) in a spatial environment. Particularly this model is useful for testing variations in T cell immunotherapies and we have integrated with multiplexed imaging datasets for initialization of the model and also verifying expected behavior.
Notebook Tutorials
Static notebooks:
- Model Tutorial: Introduces the model, including the different functions and their interactions.
- Killing Experiments: A notebook for modeling in-vitro killing experiments that can be run from a machine with the repository installed locally.
Colab notebooks:
This Model Tutorial can be altered by the user in a online environment and used to run experiments:
Description of models
Processes
The model is composed of two major cell types (T cells & Tumor cells), each with two separate phenotpyes. Each cell has an associated Vivarium Process. The process for each cell type contains the fundamental rules that govern its behavior in interacting with the other cell types and with the inputs it receives from the environment.
Testing the processes individually enables understanding if underlying parameters derived from literature values or primary data accurately represent behavior expected based on such research.
Tumor process
The tumor process is focused on two states of a tumor: proliferative with low levels of immune molecules
(MHCI and PDL1) and quiescent with high levels of immune molecules (MHCI and PDL1). Its transition from the
proliferative state is dependent on the level of interferon gamma it is exposed to coming from the T cells.
Both tumor types can be killed by recieving cytotoxic packets from the T cells.
T cell process
The T cell process is focused on two states of a T cell: PD1- with increased secretion of immune molecules
(IFNg and cytotoxic packets) and PD1+ with decreased secretion of immune molecules (IFNg and cytotoxic packets).
These immune molecules have impact of the state and death of tumor cells. Its transition from the PD1- state is
dependent on the length of time it is engaged with tumor cells.
Composites
Vivarium Composites are integrated models with multiple initialized processes, and whose inter-connections are specified by a topology. The T-cell and Tumor processes shown individually above are here combined with additional processes to create T-cell and Tumor agents. These include a division process, which waits for the division flag and then carries out division; a death process, which waits for a death flag and then removes the agent; and a local field, which interfaces the external environment to support uptake and secretion for each individual agent.
Tumor Microenvironment
TumorMicroEnvironment is a composite model that simulates a 2D environment with agents that can move around in space,
exchange molecules with their neighbors, and exchange molecules with a molecular field. This composite includes a
neighbors process, which tracks the locations of individual agents and handles their exchanges, and a diffusion
process, which operates on the molecular fields.
Simulation Experiments
In this multi-scale agent-based model, the T cells can interact with tumor cells in the following ways:
- T cell receptor (TCR on T cells) and Major histocompatibility complex I receptor (MHCI on tumor cells) for activation of T cells, induction of IFNg and cytotoxic packet secretion, and slowing of T cell migration
- PD1 receptor (on T cells) and PDL1 receptor (on tumor cells) that can inhibit cell activation and induce apoptosis
- T cells secrete IFNg which tumor cells uptake and causes state switch to upregulate MHCI, PDL1, and decrease proliferation

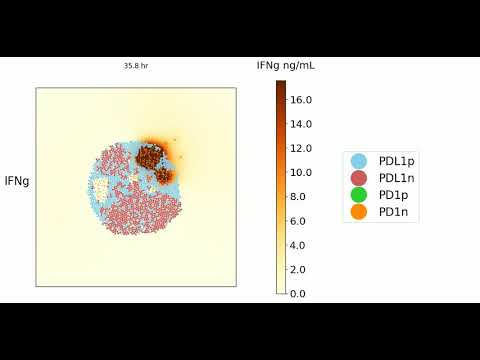
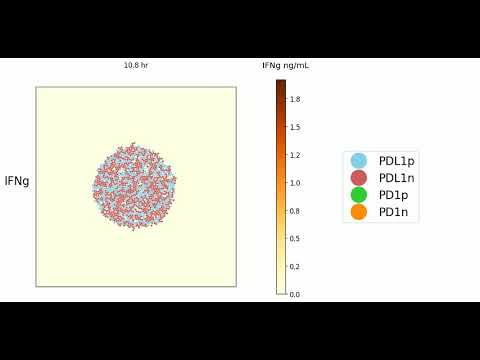
Experimental Output (click images to play on youtube)
Here are a few videos (can be generated autmomatically when running the code) that highlight simulation results initialized with 1200 cells with either no T cells or 12 T cells of varying phenotypes ran over 3 "simulation" days
25% PD1+ T cells
75% PD1+ T cells
No T cell condition
Installation
Install as Python library
Tumor-Tcell can be used as a Python library, which allows users to import modules into a different python environment, for example in a Python notebook. To install:
$ pip install tumor-tcell
Getting Started for Developers
We recommend the Vivarium Documentation.
To set up the repository for development, we recommend you clone the github repository and build a local development environment with pyenv. Pyenv lets you install and switch between multiple Python releases and multiple "virtual environments". Using pyenv, create a virtual environment and install Python 3.8.5. Follow the instructions here. Then, run the following command in your terminal:
$ pyenv virtualenv 3.8.5 tumor-tcell-env && pyenv local tumor-tcell-env
To set up the library in your environment run:
$ pip install -r requirements.txt
Set up MongoDB
We use a MongoDB database to store the data collected from running
simulations. This can be a remote server, but for this guide we will
run a MongoDB server locally. MongoDB is only required if you want to
use the database emitter in your experiments, which stores data in
MongoDB.
See Getting Started for Developers for instructions on setting up MongoDB
Running the model
Simulation experiments are specified in the file tumor_tcell/experiments/main.py.
In this file, tumor_tcell_abm is the main function for generating and simulating tumor/T cell interactions in a
2D microenvironment.
Experiments can be triggered from the command line:
$ python tumor_tcell/experiments/main.py -w [workflow id]
workflow ids are specified by the workflow_library at the bottom of the file. Workflow 3 is medium-sized
simulation that can be executed with this command:
$ python tumor_tcell/experiments/main.py -w 3
Individual processes and composites can also be executed from the command line by running their files directly. For example, run the t-cell process like this:
$ python tumor_tcell/processes/t_cell.py [--single, -s] [--batch, -b] [--timeline, -t]
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
File details
Details for the file tumor-tcell-0.0.42.tar.gz.
File metadata
- Download URL: tumor-tcell-0.0.42.tar.gz
- Upload date:
- Size: 65.2 kB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/4.0.2 CPython/3.11.4
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 7054b70bc775c86c56fdc95d8ea3606f937cd314df5fe73df4fb5a109edd6972 |
|
| MD5 | 51e41bcdcff9268faef75728d1e8d788 |
|
| BLAKE2b-256 | 022fc8e82b90d2a884eeb4e001585761afdd3a5f4f60db4af7c55775943c5a8b |