ParaDigMa - A toolbox for deriving Parkinson's disease Digital Markers from real-life wrist sensor data
Project description

| Badges | |
|---|---|
| Packages and Releases |    |
| DOI |  |
| Build Status |    |
| License |  |
Overview
ParaDigMa (Parkinson's disease Digital Markers) is a Python toolbox for extracting validated digital biomarkers from wrist sensor data in Parkinson's disease. It processes accelerometer, gyroscope, and PPG signals collected during passive monitoring in daily life.
Key Features:
- Arm swing during gait analysis
- Tremor analysis
- Pulse rate analysis
- Scientifically validated in peer-reviewed publications
- Modular, extensible architecture for custom analyses
Quick Start
Installation
For regular use:
pip install paradigma
Requires Python 3.11+.
For development or running tutorials:
Example data requires git-lfs. See the installation guide for setup instructions.
Basic Usage
from paradigma.orchestrator import run_paradigma
# Example 1: Single DataFrame with default output directory
results = run_paradigma(
dfs=df,
pipelines=['gait', 'tremor'],
watch_side='left', # Required for gait pipeline
save_intermediate=['quantification', 'aggregation'] # Saves to ./output by default
)
# Example 2: Multiple DataFrames as list (assigned to 'df_1', 'df_2', etc.)
results = run_paradigma(
dfs=[df1, df2, df3],
pipelines=['gait', 'tremor'],
output_dir="./results", # Custom output directory
watch_side='left',
save_intermediate=['quantification', 'aggregation']
)
# Example 3: Dictionary of DataFrames (custom segment/file names)
results = run_paradigma(
dfs={'morning_session': df1, 'evening_session': df2},
pipelines=['gait', 'tremor'],
watch_side='right',
save_intermediate=[] # No files saved - results only in memory
)
# Example 4: Load from data directory
results = run_paradigma(
data_path='./my_data',
pipelines=['gait', 'tremor'],
watch_side='left',
file_pattern='*.parquet',
save_intermediate=['quantification', 'aggregation']
)
# Access results (nested by pipeline)
gait_measures = results['quantifications']['gait']
tremor_measures = results['quantifications']['tremor']
gait_aggregates = results['aggregations']['gait']
tremor_aggregates = results['aggregations']['tremor']
# Check for errors
if results['errors']:
print(f"Warning: {len(results['errors'])} error(s) occurred")
See our tutorials for complete examples.
Pipelines
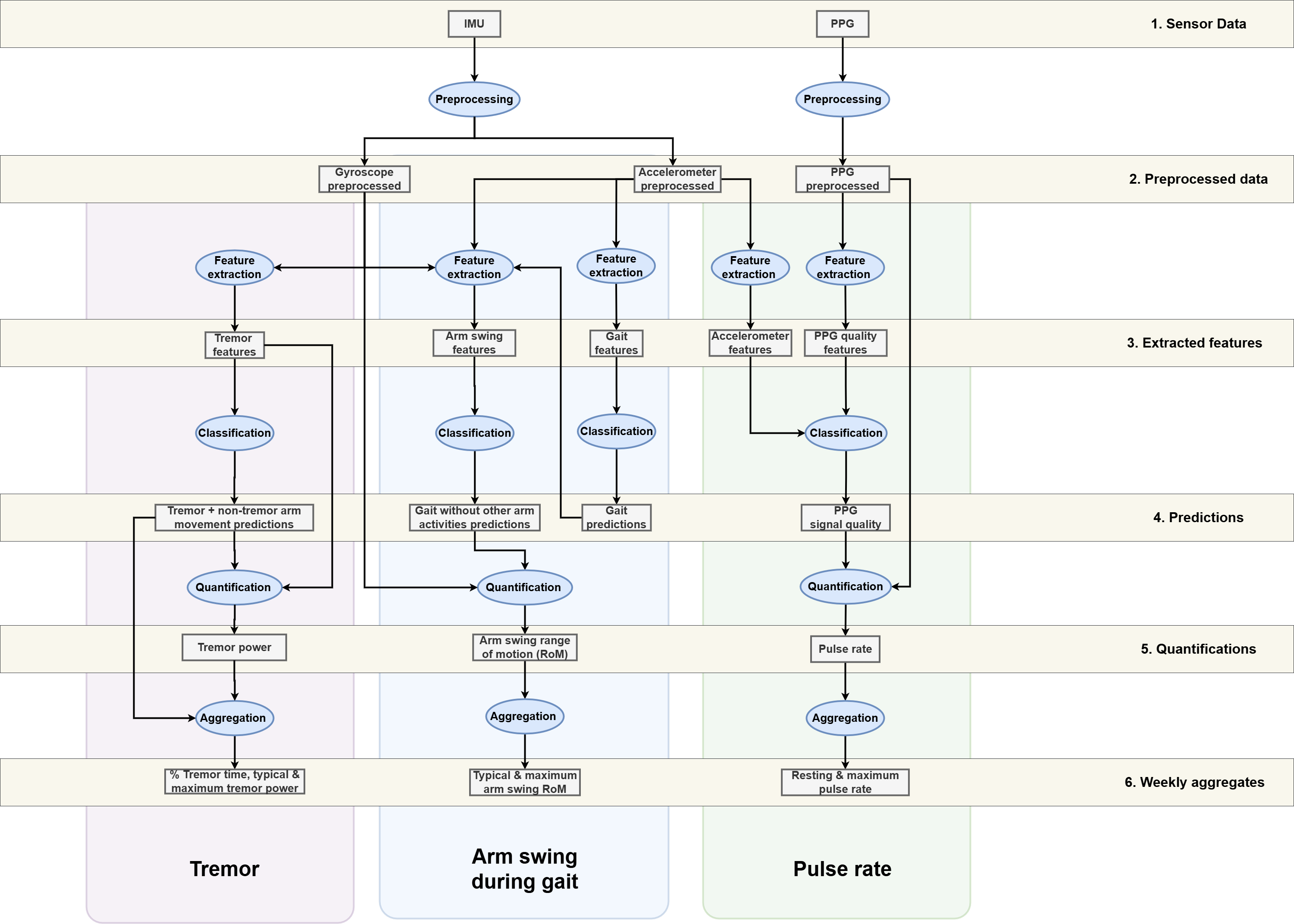
Validated Processing Pipelines
| Pipeline | Input sensors | Output week-level aggregation | Publications | Tutorial |
|---|---|---|---|---|
| Arm swing during gait | Accelerometer + Gyroscope | Typical, maximum & variability of arm swing range of motion | Post 2025, Post 2026* | Guide |
| Tremor | Gyroscope | % tremor time, typical & maximum tremor power | Timmermans 2025a, Timmermans 2025b* | Guide |
| Pulse rate | PPG (+ Accelerometer) | Resting & maximum pulse rate | Veldkamp 2025* | Guide |
* Indicates pre-print
Pipeline Architecture
ParaDigMa can best be understood by categorizing the sequential processes:
| Process | Description |
|---|---|
| Preprocessing | Preparing raw sensor signals for further processing |
| Feature extraction | Extracting features based on windowed sensor signals |
| Classification | Detecting segments of interest using validated classifiers (e.g., gait segments) |
| Quantification | Extracting specific measures from the detected segments (e.g., arm swing measures) |
| Aggregation | Aggregating the measures over a specific time period (e.g., week-level aggregates) |
Usage
Documentation
- Tutorials - Step-by-step usage examples
- Installation Guide - Setup and troubleshooting
- Sensor Requirements - Data specifications and compliance
- Supported Devices - Validated hardware
- Input Formats Guide - Input format options and data loading
- Configuration Guide - Pipeline configuration
- Scientific Validation - Validation studies and publications
- API Reference - Complete API documentation
Sensor Requirements & Supported Devices
ParaDigMa is designed for wrist sensor data collected during passive monitoring in persons with Parkinson's disease. While designed to work with any compliant device, it has been empirically validated on:
- Verily Study Watch (gait, tremor, pulse rate)
- Axivity AX6 (gait, tremor)
- Gait-up Physilog 4 (gait, tremor)
- Empatica EmbracePlus (data loading)
Please check before running the pipelines whether your sensor data complies with the requirements for the sensor configuration and context of use. See the sensor requirements guide for data specifications and the supported devices guide for device-specific setup instructions.
Data Formats
ParaDigMa supports the following data formats:
- In-memory (recommended): Pandas DataFrames (see examples above)
- Data loading file extensions: TSDF, Parquet, CSV, Pickle and several device-specific formats (AVRO (Empatica), CWA (Axivity))
Troubleshooting
For installation issues, see the installation guide troubleshooting section.
For other issues, check our issue tracker or contact paradigma@radboudumc.nl.
Scientific Validation
ParaDigMa pipelines are validated in peer-reviewed publications:
| Pipeline | Publication |
|---|---|
| Arm swing during gait | Post et al. (2025, 2026) |
| Tremor | Timmermans et al. (2025a, 2025b) |
| Pulse rate | Veldkamp et al. (2025) |
See the validation guide for full publication details.
Contributing
We welcome contributions! Please see:
Citation
If you use ParaDigMa in your research, please cite:
@software{paradigma2024,
author = {Post, Erik and Veldkamp, Kars and Timmermans, Nienke and
Soriano, Diogo Coutinho and Kasalica, Vedran and
Kok, Peter and Evers, Luc},
title = {ParaDigMa: Parkinson's disease Digital Markers},
year = {2024},
doi = {10.5281/zenodo.13838392},
url = {https://github.com/biomarkersParkinson/paradigma}
}
License
Licensed under the Apache License 2.0. See LICENSE for details.
Acknowledgements
Core Team: Erik Post, Kars Veldkamp, Nienke Timmermans, Diogo Coutinho Soriano, Vedran Kasalica, Peter Kok, Twan van Laarhoven, Luc Evers
Advisors: Max Little, Jordan Raykov, Hayriye Cagnan, Bas Bloem
Funding: the initial release was funded by the Michael J Fox Foundation (grant #020425) and the Dutch Research Council (grants #ASDI.2020.060, #2023.010)
Contact
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
Filter files by name, interpreter, ABI, and platform.
If you're not sure about the file name format, learn more about wheel file names.
Copy a direct link to the current filters
File details
Details for the file paradigma-1.1.0.tar.gz.
File metadata
- Download URL: paradigma-1.1.0.tar.gz
- Upload date:
- Size: 4.4 MB
- Tags: Source
- Uploaded using Trusted Publishing? Yes
- Uploaded via: twine/6.1.0 CPython/3.13.7
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
31c1bf47dab8dd16ce93a58ef4d6b13db3931f12e59b1ea310aa25b6b9b00343
|
|
| MD5 |
dc19c553573a210eb9e0f071a5d88ef5
|
|
| BLAKE2b-256 |
90859cc4b6f099320bff36bef5c22f51bb3f5924b01b7aab8f0559b614fcfb69
|
Provenance
The following attestation bundles were made for paradigma-1.1.0.tar.gz:
Publisher:
publish.yml on biomarkersParkinson/paradigma
-
Statement:
-
Statement type:
https://in-toto.io/Statement/v1 -
Predicate type:
https://docs.pypi.org/attestations/publish/v1 -
Subject name:
paradigma-1.1.0.tar.gz -
Subject digest:
31c1bf47dab8dd16ce93a58ef4d6b13db3931f12e59b1ea310aa25b6b9b00343 - Sigstore transparency entry: 919523206
- Sigstore integration time:
-
Permalink:
biomarkersParkinson/paradigma@e409e47551c6630da84b15cd1b6ff399feafb03a -
Branch / Tag:
refs/heads/release - Owner: https://github.com/biomarkersParkinson
-
Access:
public
-
Token Issuer:
https://token.actions.githubusercontent.com -
Runner Environment:
github-hosted -
Publication workflow:
publish.yml@e409e47551c6630da84b15cd1b6ff399feafb03a -
Trigger Event:
push
-
Statement type:
File details
Details for the file paradigma-1.1.0-py3-none-any.whl.
File metadata
- Download URL: paradigma-1.1.0-py3-none-any.whl
- Upload date:
- Size: 4.5 MB
- Tags: Python 3
- Uploaded using Trusted Publishing? Yes
- Uploaded via: twine/6.1.0 CPython/3.13.7
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
04454aabbe4c97a226a2785b9f1216a427c692f3af6b9af832ab1fc3f151dd7d
|
|
| MD5 |
b8b9022d1faedd827149fd8ee7d29508
|
|
| BLAKE2b-256 |
ae712e76e82ad3a2f0d57fcca5721c55d856be29ee2744e48cf6fc2d9ccd3234
|
Provenance
The following attestation bundles were made for paradigma-1.1.0-py3-none-any.whl:
Publisher:
publish.yml on biomarkersParkinson/paradigma
-
Statement:
-
Statement type:
https://in-toto.io/Statement/v1 -
Predicate type:
https://docs.pypi.org/attestations/publish/v1 -
Subject name:
paradigma-1.1.0-py3-none-any.whl -
Subject digest:
04454aabbe4c97a226a2785b9f1216a427c692f3af6b9af832ab1fc3f151dd7d - Sigstore transparency entry: 919523216
- Sigstore integration time:
-
Permalink:
biomarkersParkinson/paradigma@e409e47551c6630da84b15cd1b6ff399feafb03a -
Branch / Tag:
refs/heads/release - Owner: https://github.com/biomarkersParkinson
-
Access:
public
-
Token Issuer:
https://token.actions.githubusercontent.com -
Runner Environment:
github-hosted -
Publication workflow:
publish.yml@e409e47551c6630da84b15cd1b6ff399feafb03a -
Trigger Event:
push
-
Statement type:















