A graph benchmark library for heterophilic and heterogeneous graphs
Project description

Overview
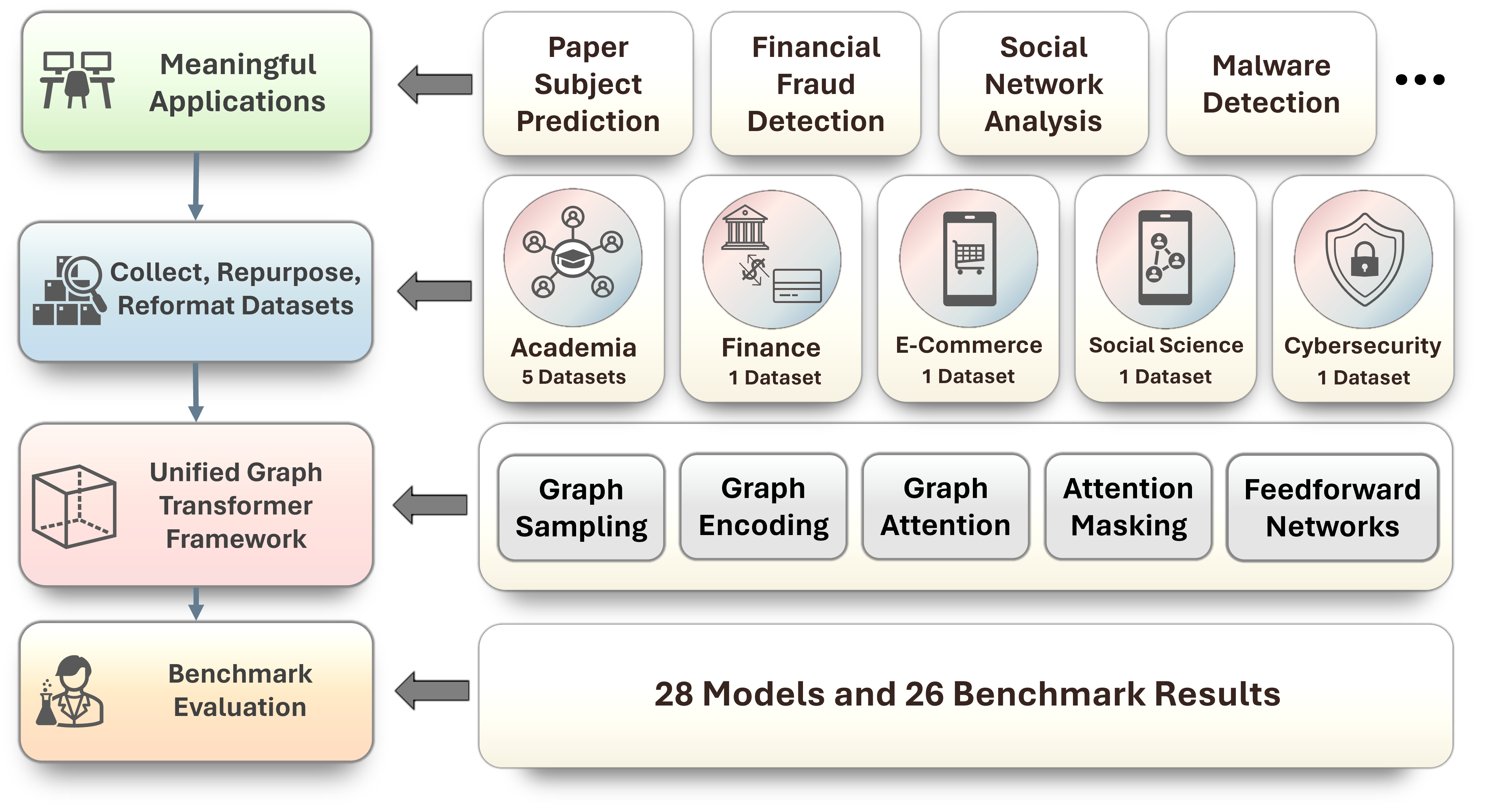
The Heterophilic and Heterogeneous Graph Benchmark (H²GB) is a collection of graph benchmark datasets, data loaders, modular graph transformer framework (UnifiedGT) and evaluators for graph learning. The H²GB encompasses 9 diverse real-world datasets across 5 domains. Its data loaders are fully compatible with popular graph deep learning framework PyTorch Geometric. They provide automatic dataset downloading, standardized dataset splits, and unified performance evaluation.
Environment Setup
You can create a conda environment to easily run the code. For example, we can create a virtual environment named H2GB:
conda create -n H2GB python=3.9 -y
conda activate H2GB
Install the required packages using the following commands:
conda install pytorch torchvision torchaudio pytorch-cuda=11.8 -c pytorch -c nvidia
conda install pyg -c pyg
pip install -r requirements.txt
Run the UnifiedGT
To summarize and systematically compare the performance of existing GNNs on H2GB, we designed UnifiedGT. UnifiedGT is a modular graph transformer framework that designed to encompass many existing GTs and GNNs by leveraging unified components: (1) graph sampling, (2) graph encoding, (3) graph attention, (4) attention masking, and (5) feedforward networks (FFN). It is implemented as a Python library and is user-friendly. It includes a unified data loader and evaluator, making it easy for researchers to access datasets, evaluate methods, and compare performance.
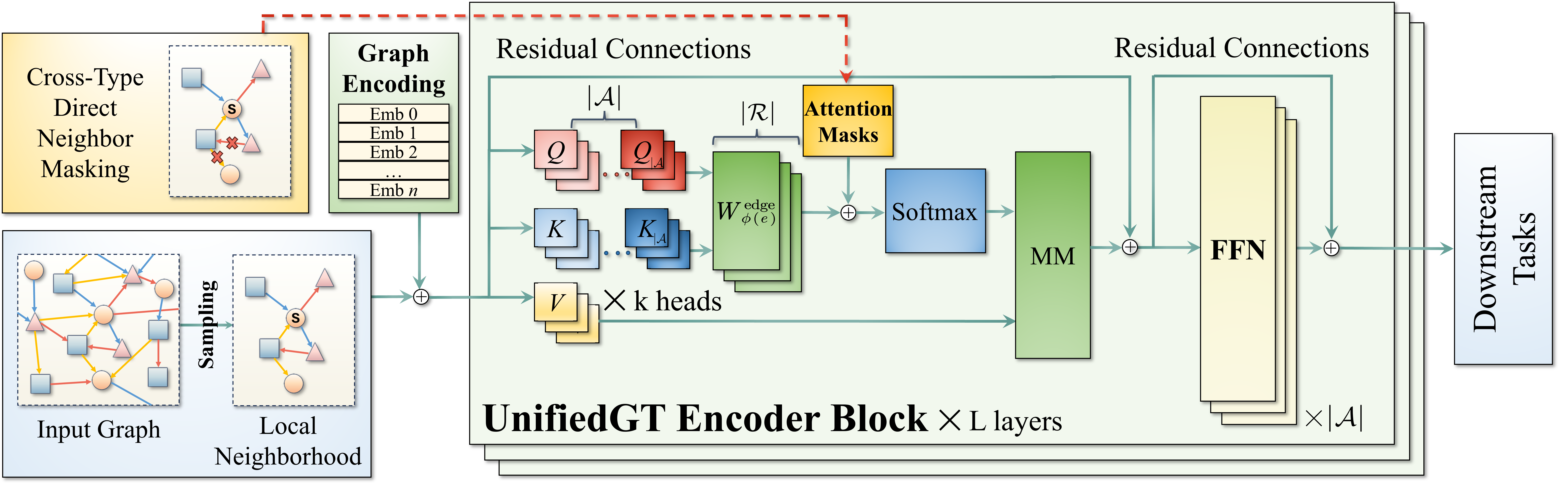
We implement 9 existing GT baselines and 19 GNN models based on UnifiedGT and provide comprehensive experiment configurations available in ./configs. To run UnifiedGT, you will need to firstly specify the dataset and log location by editing the config file provided under ./configs/{dataset_name}/. An example configuration is
......
out_dir: ./results/{Model Name} # Put your log output path here
dataset:
dir: ./data # Put your input data path here
......
Dataset download will be automatically initiated if dataset is not found under the specified location.
For convenience, a script file is created to run the experiment with specified configuration. For instance, you can edit and run the interactive_run.sh to start the experiment.
# Assuming you are located in the H2GB repo
chmox +x ./run/interactive_run.sh
./run/interactive_run.sh
You can also directly enter this command into your terminal:
python -m H2GB.main --cfg {Path to Your Configs} name_tag {Custom Name Tag}
For example, the following command is to run MLP model experiment for oag-cs dataset.
python -m H2GB.main --cfg configs/oag-cs/oag-cs-MLP.yaml name_tag MLP
Caclulate the Metapath-Induced Adjusted Heterophily Measurement
We provide a extended heterophily measurement from homogeneous grpah into the heterogeneous setting, which is called metapath-induced heterophily measrement. The calcualtion function is available in ./H2GB/calcHomophily.py. You can simply import it by using from H2GB.calcHomophily import calcHomophily and measure the heterophily of your data. For convenience, we also provide a script to reproduce the heterophily measurement on our developed datasets. Note that $$\text{Heterophily} = 1 - \text{Homophily}$$ So just do a simple transformation to obtain the heterophily.
chmox +x ./run/calcHomo.sh
./run/calcHomo.sh
Side Notes
Encoders
The Hetero_Raw encoder are supposed to be used for heterogeneous GNN or graph dataset that has different node encoding dimensions for different node type. Therefore, each node type can be transformed separately. To reproduce results of homogeneous GNN, consider using the Raw encoder, which apply the same transformation for each node type. Otherwise, using Hetero_Raw for homogeneous GNN will misleadingly increase the task performance.
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.














