A desktop platform for streamlined molecular sequence data management and state of the art evolutionary phylogenetics studies.
Project description
PhyloSuite
PhyloSuite is an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. It combines the functions of two previous tools: MitoTool (https://github.com/dongzhang0725/MitoTool) and BioSuite (https://github.com/dongzhang0725/BioSuite).
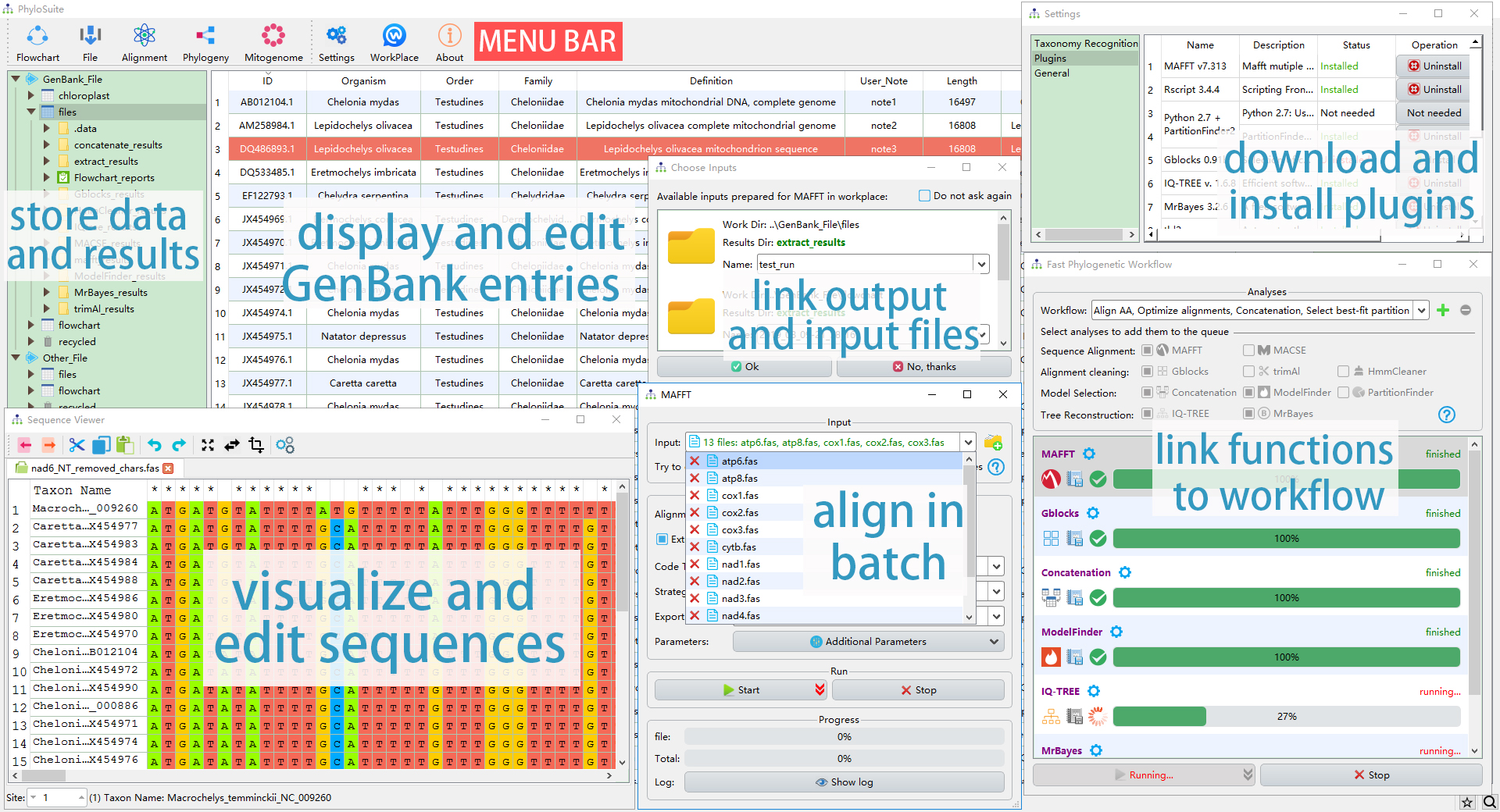
The interface and the main functions of PhyloSuite.
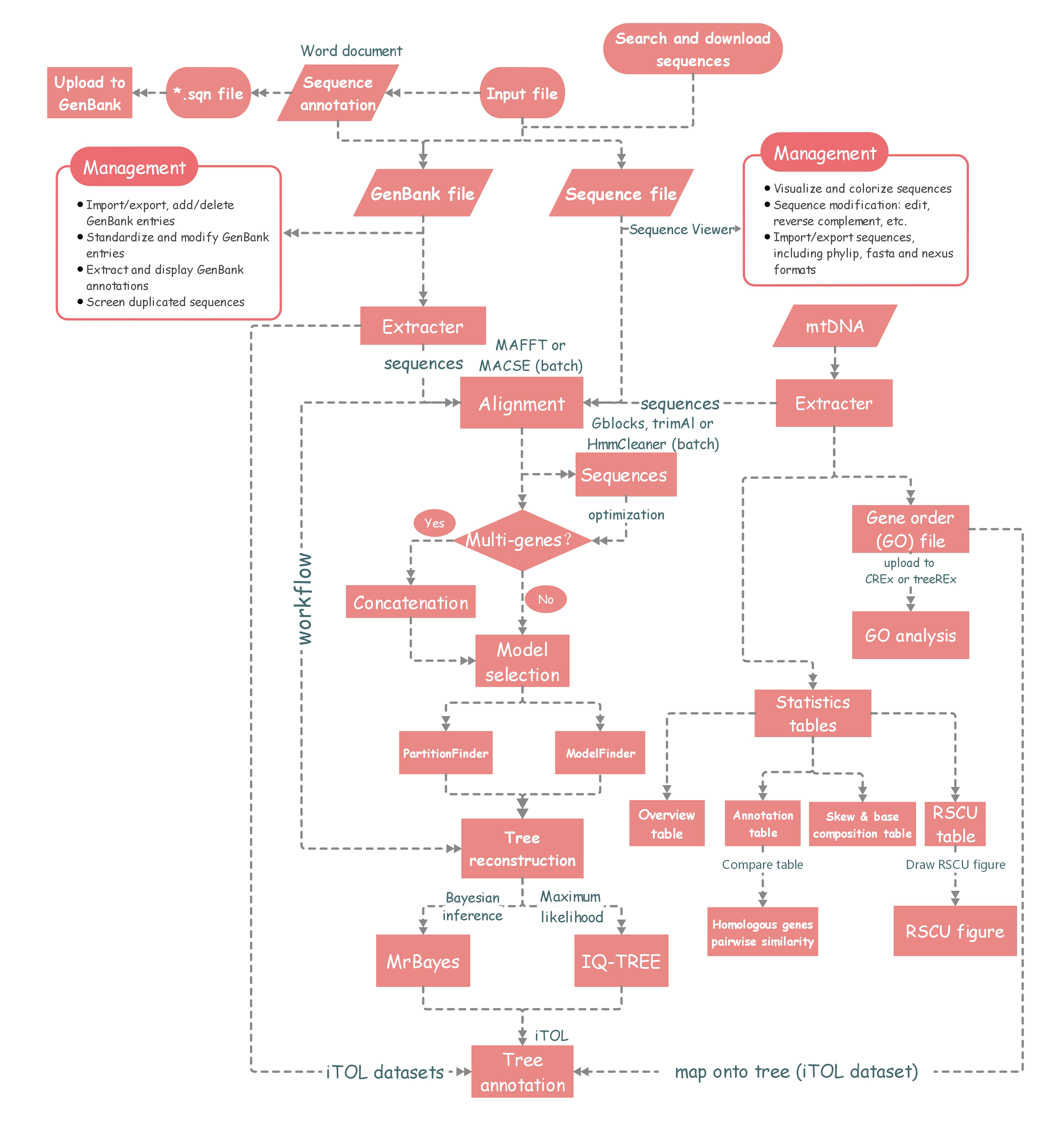
The workflow diagram of PhyloSuite.
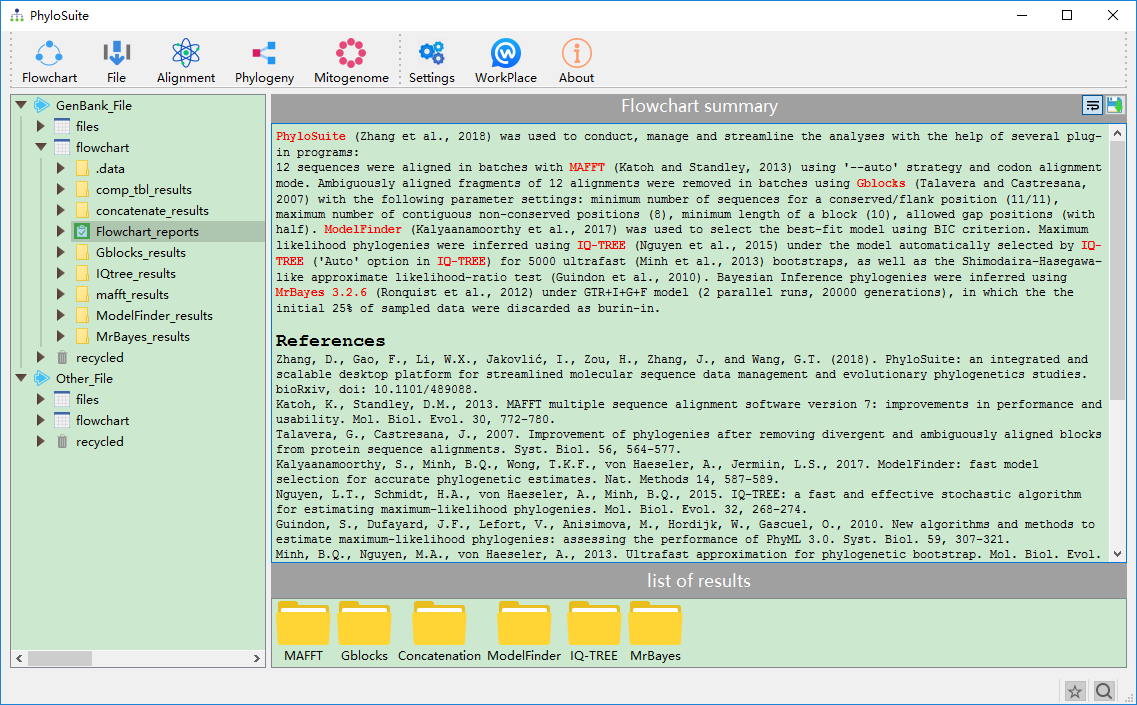
The summary of flowchart and references of used programs.
Homepage
https://dongzhang0725.github.io
Installation
1. Install compiled PhyloSuite
Installers for all platforms can be downloaded from https://github.com/dongzhang0725/PhyloSuite/releases.
1.1. Windows
Windows 7, 8 and 10 are supported, just double click the PhyloSuite_xxx_win_setup.exe to install, and run “PhyloSuite.exe” file after the installation. If the installation fails, download PhyloSuite_xxx_Win.rar, unzip it, and run PhyloSuite directly from this folder.
1.2. Mac OSX & Linux
Unzip PhyloSuite_xxx_Mac.zip/PhyloSuite_xxx_Linux.tar.gz to anywhere you like, and double click “PhyloSuite” (in PhyloSuite folder) to start, or use the following command:
cd path/to/PhyloSuite
./PhyloSuite
If you encounter an error of "permission denied", try to use the following command:
chmod -R 755 path/to/PhyloSuite(folder)
Note that both 64 bit and 32 bit Windows is supported (Windows 7 and above), whereas only 64 bit has been tested in Linux (Ubuntu 14.04.1 and above) and Mac OSX (macOS Sierra version 10.12.3 and above).
2. Install using pip
First, Python (version higher than 3.6) should be installed and added to the environment variable in your computer. Then open the terminal and type:
pip install PhyloSuite
It will take some time to install. If it installs successfully, PhyloSuite will be automatically added to the environment variables. Then open the terminal again and type:
PhyloSuite
If the above pip command failed to install PhyloSuite, you can use compiled PhyloSuite (see section 1) or find and download the source codes here (https://pypi.org/project/PhyloSuite/#files or https://github.com/dongzhang0725/PhyloSuite), and install it manually.
Examples
https://raw.githubusercontent.com/dongzhang0725/PhyloSuite/master/example.zip
Bug report
https://github.com/dongzhang0725/PhyloSuite/issues or send email to dongzhang0725@gmail.com.
Citation
Zhang, D., Gao, F., Li, W.X., Jakovlić, I., Zou, H., Zhang, J., and Wang, G.T. (2018). PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. bioRxiv, doi: 10.1101/489088. (Download as: RIS XML ENW)
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
Hashes for PhyloSuite-1.1.16.post1-py2.py3-none-any.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 04a4d071c0c7692639fce6db9b8ea55c9b8fbe50ec7725dc81a892e0444eae44 |
|
| MD5 | 151fb230a097b588655ef2b05cd58270 |
|
| BLAKE2b-256 | b3905141ae526bdea481b6c98adb426df7f25d916c0c193370d82dcb1466f568 |