A high performance mapping class to construct ElM2D plots from large datasets of inorganic compositions.
Project description
chem_wasserstein
A high performance mapping class to construct ElMD distance matrices from large datasets of ionic compositions, suitable for single node usage on HPC systems. This includes helper methods to directly embed these datasets as maps of chemical space, as well as sorting lists of compositions, and exporting kernel matrices.
This Fork
:warning: For the original ElM2D repository, see https://github.com/lrcfmd/ElM2D. :warning:
This is a refactored version which incorporates fast distance matrix computations for the modified Pettifor scale representation, and is in the process of being integrated into ElMD and ElM2D. The documentation as follows has changes relative to the original documentation. Additionally, this is packaged on PyPI and Anaconda, but under a different name: chem_wasserstein. A $10000 \times 10000$ pairwise distance matrix can be calculated on the order of 10 seconds (CPU). The distance calculations are also GPU compatible. If running on Colab, be sure to use GPU as the CPU version has some Colab-specific issues with Numba.
Installation
Recommended installation through conda with python 3.8.
conda install -c sgbaird chem_wasserstein
or
pip install chem_wasserstein
For the background theory, please read the paper "The Earth Mover’s Distance as a Metric for the Space of Inorganic Compositions"
Examples
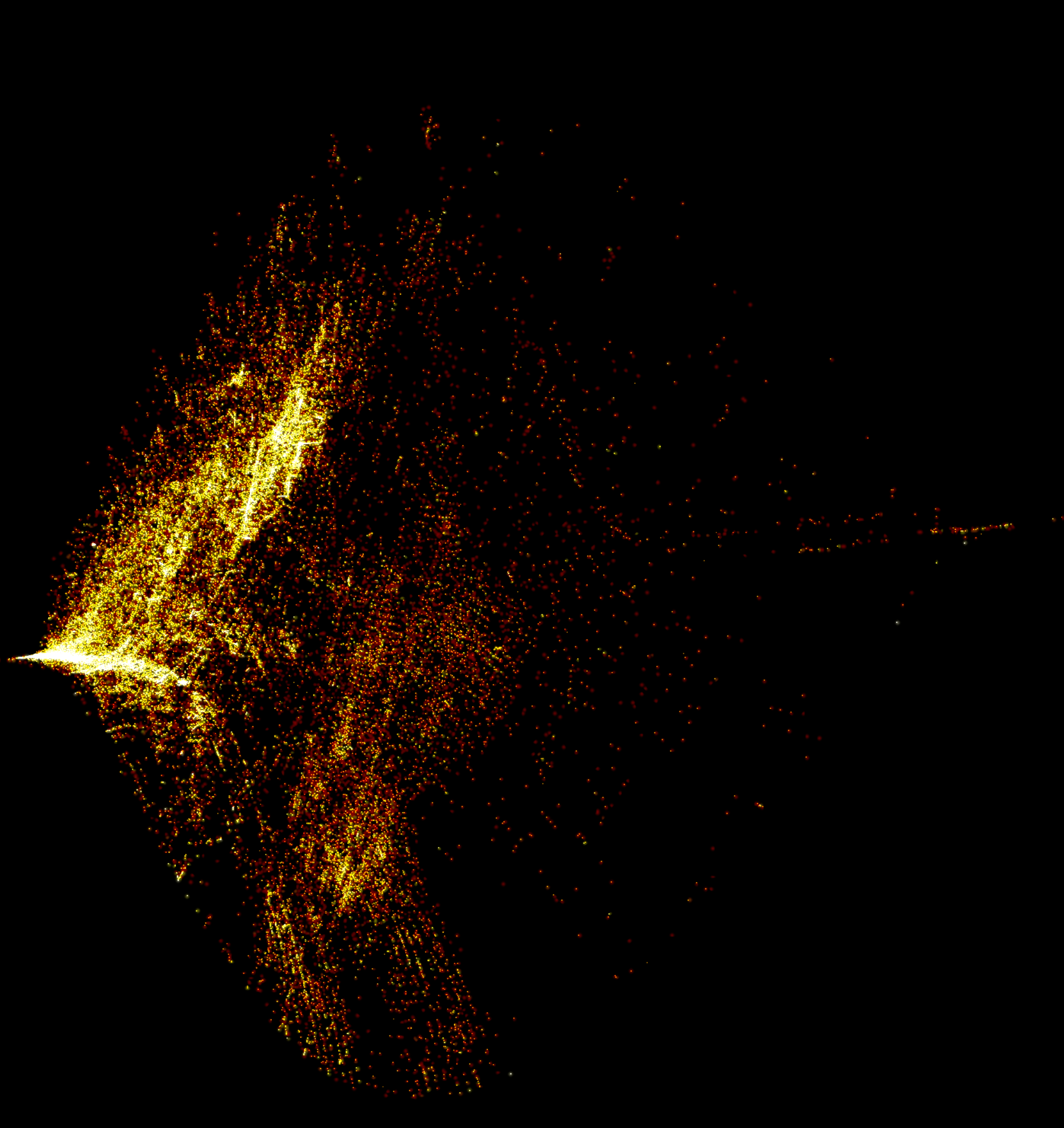
125,000 compositions from the inorganic crystal structure database embedded with PCA, plotted with datashader:
For more interactive examples please see www.elmd.io/plots
Usage
Computing Distance Matrices
The computed distance matrix is accessible through the dm attribute and can be saved and loaded as a csv file.
from chem_wasserstein.ElM2D_ import ElM2D
mapper = ElM2D()
mapper.fit(df["formula"])
print(mapper.dm)
mapper.export_dm("ComputedMatrix.csv")
This distance matrix can be used as a lookup table for distances between compositions given their numeric indices (distance = mapper.dm[i][j]) or used as a kernel matrix for embedding, regression, and classification tasks directly.
Sorting
To sort a list of compositions into an ordering of chemical similarity
mapper.fit(df["formula"])
sorted_indices = mapper.sort()
sorted_comps = mapper.sorted_formulas
Embedding
Embeddings can be constructed through either the UMAP or PCA methods of dimensionality reduction. The most recently embedded points are accessible via the embedding property. Higher dimensional embeddings can be created with the n_components parameter.
mapper = ElM2D()
mapper.fit(df["formula"])
embedding = mapper.transform()
...
# For new data
embedding = mapper.fit_transform(df["formula"])
embedding = mapper.fit_transform(df["formula"], how="PCA", n_components=7)
Embeddings may also be directed towards a particular chemical property in a pandas DataFrame, to bring known patterns into focus.
embedding = mapper.fit_transform(df["formula"], df["property_of_interest"])
By default, the modified Pettifor scale is used as the method of atomic similarity, this is changed through the metric attribute.
mapper = ElM2D(metric="atomic")
embedding = mapper.fit_transform(df["formula"])
These embeddings may be visualized within a jupyter notebook, or exported to HTML to view full page in the web browser.
mapper.fit_transform(df["formula"])
# Returns a figure for viewing in notebooks
mapper.plot()
# Returns a figure and saves as ElM2D_Plot_UMAP.html
mapper.plot("ElM2D_Plot_UMAP.html")
# Returns and saves figure, with colouring based on property from a pandas Series
mapper.plot(fp="ElM2D_Plot_UMAP.html", color=df["chemical_property"])
# Plotting also works in 3D
mapper.fit_transform(df["formula"], n_components=3)
mapper.plot(color=df["chemical_property"])
Saving
Smaller datasets can be saved directly with the save(filepath.pk)/load(filepath.pk) methods directly. This is limited to files of size 3GB (the python binary file size limit).
Larger datasets will require importing/exporting the distance matrix and embeddings (export_embedding(filepath.csv)/import_embedding(filepath.csv) separately as csv files if you require this processed data in future work.
mapper.fit(small_df["formula"])
mapper.save("small_df_mapper.pk")
...
mapper = ElM2D()
mapper.load("small_df_mapper.pk")
...
mapper.fit(large_df["formula"])
mapper.export_dm("large_df_dm.csv")
mapper.export_embedding("large_df_emb_UMAP.csv")
...
mapper = ElM2D()
mapper.import_dm("large_df_dm.csv")
mapper.import_embedding("large_df_emb_UMAP.csv")
mapper.formula_list = df["formula"]
Cross Validation
Perform a K-Folds splitting of the dataset into subsets, to build up training and testing datasets.
cvs = mapper.cross_validate()
for i, (X_train, X_test) in enumerate(cvs):
sub_mapper = ElM2D()
sub_mapper.fit(X_train)
sub_mapper.save(f"train_elm2d_{i}.pk")
sub_mapper.fit(X_test)
sub_mapper.save(f"test_elm2d_{i}.pk")
...
from sklearn.metrics import mean_average_error as mae
cvs = mapper.cross_validate(y=df["target"])
for X_train, X_test, y_train, y_test in cvs:
clf.fit(X_train, y_train)
y_pred = clf.predict(X_test)
errors.append(mae(y_pred, y_test))
print(np.mean(errors))
Available Metrics
You may use either discrete scales or machine learnt representations for each element. Choose these via the metric parameter.
Linear:
- mendeleev
- petti
- atomic
- mod_petti
Chemically Derived:
- oliynyk
- oliynyk_sc
- jarvis
- jarvis_sc
- magpie
- magpie_sc
Machine Learnt:
- cgcnn
- elemnet
- mat2vec
- matscholar
- megnet16
Random Numbers:
- random_200
Custom Distance Matrix
- precomputed
Bulk featurizing can be performed with featurize.
mapper = ElM2D(metric="oliynyk_sc")
X = mapper.featurize(df["formula"])
Citing
If you would like to cite this code in your work, please use the following reference
@article{doi:10.1021/acs.chemmater.0c03381,
author = {Hargreaves, Cameron J. and Dyer, Matthew S. and Gaultois, Michael W. and Kurlin, Vitaliy A. and Rosseinsky, Matthew J.},
title = {The Earth Mover’s Distance as a Metric for the Space of Inorganic Compositions},
journal = {Chemistry of Materials},
volume = {32},
number = {24},
pages = {10610-10620},
year = {2020},
doi = {10.1021/acs.chemmater.0c03381},
URL = {
https://doi.org/10.1021/acs.chemmater.0c03381
},
eprint = {
https://doi.org/10.1021/acs.chemmater.0c03381
}
}
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
File details
Details for the file chem_wasserstein-1.0.12.tar.gz.
File metadata
- Download URL: chem_wasserstein-1.0.12.tar.gz
- Upload date:
- Size: 1.0 MB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: python-requests/2.31.0
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | b12f7772a4ed075d11ce7768682bf70c34db39867d06aa8bbae5ad5ff01b901e |
|
| MD5 | 5f6b2f2cc22f16cbf0b8f6ec4aeb1540 |
|
| BLAKE2b-256 | 6f6af4d6a3945ddbd51379fd0cb5ed6c8edc6c866e76fecc015d42abd4225a72 |
File details
Details for the file chem_wasserstein-1.0.12-py3-none-any.whl.
File metadata
- Download URL: chem_wasserstein-1.0.12-py3-none-any.whl
- Upload date:
- Size: 1.0 MB
- Tags: Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: python-requests/2.31.0
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | b38963667f343343067d241028450380b53fc03eb486edf980e3b08ef3258310 |
|
| MD5 | 4408b93c81556cb2dcb5ca8c66bbfe64 |
|
| BLAKE2b-256 | 899f82e9e9bf3116caddfebc8670af8c46fa6f364acea64c179bc8435c0dd0de |