An Enhanced Adaptive DBSCAN clustering algorithm for semiconductor wafer defects.
Project description
Enhanced Adaptive DBSCAN
An Enhanced Adaptive DBSCAN clustering algorithm tailored for semiconductor wafer defect analysis and other applications requiring adaptive density-based clustering.
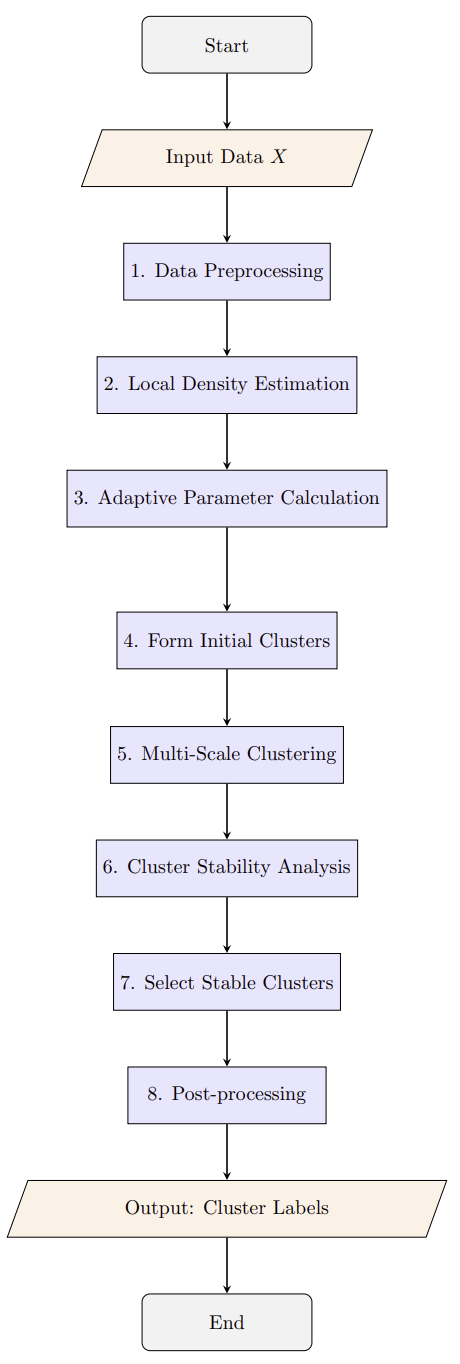
Illustration of the Enhanced Adaptive DBSCAN clustering process.
Table of Contents
Introduction
The Enhanced Adaptive DBSCAN algorithm is an advanced clustering method that extends the traditional DBSCAN algorithm by incorporating adaptive parameters and stability analysis. It is particularly designed to handle complex datasets with varying densities, such as those found in semiconductor wafer defect analysis.
Features
Adaptive Parameter Selection: Automatically adjusts ε (epsilon) and MinPts for each data point based on local density.
Stability-Based Cluster Selection: Identifies and retains robust clusters through multi-scale analysis.
Dynamic Cluster Centers: Maintains updated cluster centroids for accurate representation.
Partial Re-Clustering: Efficiently updates clusters affected by new data points without reprocessing the entire dataset.
Incremental Clustering: Supports real-time data by incrementally updating clusters with new incoming data.
Interactive Visualization: Provides interactive plots using Plotly for exploratory data analysis.
Comprehensive Logging: Offers detailed logging for monitoring the clustering process and debugging.
Installation
You can install the package using pip:
pip install enhanced_adaptive_dbscan
Quick Start
Here’s a quick example to get you started:
from enhanced_adaptive_dbscan import EnhancedAdaptiveDBSCAN
import numpy as np
# Generate synthetic data
X = np.random.randn(1000, 2)
severity = np.random.randint(1, 11, size=(1000, 1))
X_full = np.hstack((X, severity))
# Initialize the model
model = EnhancedAdaptiveDBSCAN(
wafer_shape='circular',
wafer_size=100,
k=20,
density_scaling=1.0,
buffer_ratio=0.1,
min_scaling=5,
max_scaling=10,
n_jobs=-1,
max_points=100000,
subsample_ratio=0.1,
random_state=42,
additional_features=[2], # Index of 'severity' in X_full
feature_weights=[1.0],
stability_threshold=0.6
)
# Fit the model
model.fit(X_full, additional_attributes=severity)
# Retrieve cluster labels
labels = model.labels_
# Plot the clusters
model.plot_clusters(X_full, plot_all=False)
# Evaluate clustering
model.evaluate_clustering(X_full[:, :2], labels)
Algorithm Overview
The Enhanced Adaptive DBSCAN algorithm builds upon the classic DBSCAN algorithm by introducing adaptive parameters and stability analysis to better handle datasets with varying densities.
Adaptive Parameter Selection
Local Density Estimation: Computes the local density around each data point using k-nearest neighbors.
Adaptive Epsilon (ε): Adjusts the ε parameter based on local density to capture clusters of varying densities.
Adaptive MinPts: Modifies the MinPts parameter dynamically for each point to reflect local point density.
Stability-Based Cluster Selection
Multi-Scale Clustering: Performs clustering across multiple density scales.
Stability Analysis: Evaluates clusters based on their persistence across scales, retaining only stable clusters.
Thresholding: Clusters with stability above stability_threshold are retained.
Dynamic Cluster Centers
Centroid Updates: Maintains and updates the centroids of clusters as new data points are assigned.
Representation Accuracy: Ensures that the cluster centers accurately represent the current state of the clusters.
Partial Re-Clustering
Affected Clusters Identification: Detects clusters affected by new data points.
Efficient Updates: Re-clusters only the affected clusters instead of the entire dataset, improving performance.
Incremental Clustering
Real-Time Data Handling: Supports the addition of new data points without reprocessing the entire dataset.
Incremental Updates: Updates clusters and centroids incrementally to accommodate streaming data.
API Reference
Class: EnhancedAdaptiveDBSCAN
EnhancedAdaptiveDBSCAN(
wafer_shape='circular',
wafer_size=100,
k=20,
density_scaling=1.0,
buffer_ratio=0.1,
min_scaling=5,
max_scaling=10,
n_jobs=-1,
max_points=100000,
subsample_ratio=0.1,
random_state=None,
additional_features=None,
feature_weights=None,
stability_threshold=0.6
)Parameters:
wafer_shape (str): Shape of the wafer (‘circular’ or ‘square’).
wafer_size (float): Size of the wafer (radius for circular, side length for square).
k (int): Number of neighbors for density estimation.
density_scaling (float): Scaling factor for adaptive ε.
buffer_ratio (float): Fraction of wafer size to create a buffer zone near boundaries.
min_scaling (int): Minimum scaling factor for adaptive MinPts.
max_scaling (int): Maximum scaling factor for adaptive MinPts.
n_jobs (int): Number of parallel jobs for multiprocessing (-1 uses all available cores).
max_points (int): Threshold for maximum number of points before subsampling is applied.
subsample_ratio (float): Ratio of data to subsample when max_points is exceeded.
random_state (int or None): Seed for reproducibility in subsampling.
additional_features (list or None): List of additional feature indices to include in clustering.
feature_weights (list or None): Weights for additional features to balance their influence.
stability_threshold (float): Minimum stability score to retain a cluster.
Attributes:
labels_ (ndarray): Cluster labels for each point in the dataset.
cluster_centers_ (dict): Centroids of the clusters.
cluster_stability_ (dict): Stability scores of the clusters.
Methods:
fit(X, additional_attributes=None): Fits the model to the data X.
fit_incremental(X_new, additional_attributes_new=None): Incrementally fits new data X_new to the existing model.
plot_clusters(X, plot_all=False, title=’Enhanced Adaptive DBSCAN Clustering’): Plots the clustering results.
evaluate_clustering(X, labels): Evaluates the clustering using internal metrics.
Examples
Example 1: Clustering Synthetic Data
import numpy as np
from enhanced_adaptive_dbscan import EnhancedAdaptiveDBSCAN
# Generate synthetic data with clusters of varying densities
np.random.seed(0)
cluster_1 = np.random.normal(0, 0.5, size=(100, 2))
cluster_2 = np.random.normal(5, 1.5, size=(300, 2))
cluster_3 = np.random.normal(-5, 1.0, size=(200, 2))
X = np.vstack((cluster_1, cluster_2, cluster_3))
# Initialize the model
model = EnhancedAdaptiveDBSCAN(k=15, stability_threshold=0.5)
# Fit the model
model.fit(X)
# Plot the clusters
model.plot_clusters(X)
# Evaluate clustering
model.evaluate_clustering(X, model.labels_)Example 2: Incremental Clustering
# Assume 'model' is already fitted on initial data 'X_initial'
# New incoming data
X_new = np.random.normal(0, 0.5, size=(50, 2))
# Incrementally fit new data
model.fit_incremental(X_new)
# Update labels and plot
labels_updated = model.labels_
model.plot_clusters(np.vstack((X, X_new)))Contributing
Contributions are welcome! If you’d like to contribute to this project, please follow these steps:
Fork the Repository: Click on the ‘Fork’ button on GitHub.
Clone Your Fork:
git clone https://github.com/kennedym-ds/enhanced_adaptive_dbscan.gitCreate a Branch:
git checkout -b feature/your-feature-nameMake Your Changes: Implement your feature or bug fix.
Commit Your Changes:
git commit -m "Description of your changes"Push to Your Fork:
git push origin feature/your-feature-nameSubmit a Pull Request: Go to the original repository and create a pull request.
Please ensure your code follows the project’s coding standards and includes appropriate tests.
License
This project is licensed under the MIT License.
Additional Information
Dependencies
Python >= 3.6
NumPy
SciPy
scikit-learn
Plotly
pandas
joblib
Support
If you encounter any issues or have questions, please open an issue on the GitHub repository.
Acknowledgments
The implementation is inspired by advancements in density-based clustering algorithms and their applications in semiconductor manufacturing and other industries.
Algorithm Details
Adaptive Epsilon (ε) Calculation
The ε parameter is computed individually for each data point based on local density:
Local Density Estimation: For each point, compute the average distance to its k-nearest neighbors.
Adaptive ε: ε is inversely proportional to the local density:
\begin{equation*} \epsilon_i = \frac{C}{\text{density}_i + \delta} \end{equation*}where ( C ) is a scaling constant and ( delta ) is a small value to prevent division by zero.
Adaptive MinPts Calculation
MinPts is adjusted to reflect the local point density:
Normalization: Normalize local densities to a range between min_scaling and max_scaling.
Adaptive MinPts: Assign MinPts to each point based on the normalized density.
Stability Analysis
Multi-Scale Clustering: Perform clustering at multiple scales by varying ε and MinPts within defined ranges.
Cluster Stability: A cluster’s stability is determined by its presence across multiple scales.
Thresholding: Clusters with stability above stability_threshold are retained.
Incremental Clustering
New Data Integration: New data points are assigned to existing clusters if they fall within the adaptive ε of cluster members.
Partial Re-Clustering: Only affected clusters are re-clustered when new data significantly alters the cluster structure.
Boundary Handling
Buffer Zones: A buffer zone near the wafer boundary adjusts densities to account for edge effects.
Density Adjustment: Densities near boundaries are scaled to prevent artificial inflation or deflation.
Frequently Asked Questions
Q1: How does this algorithm differ from standard DBSCAN?
The Enhanced Adaptive DBSCAN algorithm introduces adaptive parameters for ε and MinPts, allowing it to handle datasets with varying densities more effectively. It also incorporates stability analysis to retain robust clusters and supports incremental updates.
Q2: Can I use this algorithm for non-wafer datasets?
Yes, while it is tailored for semiconductor wafer defect analysis, the algorithm is generic and can be applied to any dataset where adaptive density-based clustering is beneficial.
Q3: How do I choose the parameters like ``k``, ``density_scaling``, and ``stability_threshold``?
``k``: Typically set based on the expected local neighborhood size. A higher k smooths out density estimates.
``density_scaling``: Adjusts the sensitivity of ε to local densities. Experimentation may be needed.
``stability_threshold``: Determines how persistent a cluster must be across scales to be retained. A value between 0.5 and 0.7 is common.
Citation
If you use this algorithm in your research, please consider citing:
@software{michaelKennedy_enhancedadaptiveDBSCAN_2024,
author = {Michael Kennedy},
title = {{Enhanced Adaptive DBSCAN}},
year = {2024},
url = {https://github.com/kennedym-ds/enhanced_adaptive_dbscan},
}Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
File details
Details for the file enhanced_adaptive_dbscan-0.1.1.tar.gz.
File metadata
- Download URL: enhanced_adaptive_dbscan-0.1.1.tar.gz
- Upload date:
- Size: 20.4 kB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/5.1.1 CPython/3.12.4
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | a119b099b7038de76d2c1ecceb4aeaef43315af49446145b4946c4fcb57ed91b |
|
| MD5 | 931d62ab841531e35e075eb4b2c8b5a7 |
|
| BLAKE2b-256 | 424fae08e41b462f648ecf78a1f8b05991c4979afa573b5acc92e7c50e3e51f4 |
File details
Details for the file enhanced_adaptive_dbscan-0.1.1-py3-none-any.whl.
File metadata
- Download URL: enhanced_adaptive_dbscan-0.1.1-py3-none-any.whl
- Upload date:
- Size: 15.5 kB
- Tags: Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/5.1.1 CPython/3.12.4
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 6400a736c2df9689caef24df4f6ed3822b0f6d5a7317d59380f44f4eb281be12 |
|
| MD5 | b0df01e4621822d7750cae49b5552d75 |
|
| BLAKE2b-256 | 3c6769d68cd231441c09fefdec7a199c592c47a1021c17d8c5cd7a6327af8b90 |











