A python package to quickly get and visualise the gene order/synteny around a target gene from one or more species.
Project description
EZgenesynteny
Query GenBank to obtain gene orders from genomic records and simply visualise synteny between different species.
- getgenes.py: query GenBank database with species and gene name to obtain order of surrounding protien-coding genes
- genevis.py: functions to graphically visualise the gene order/synteny and save to image file
- emailaddress.py: check and update the email address used to make GenBank Entrez queries
Author
Jake Leyhr (@jakeleyhr)
https://github.com/jakeleyhr/EZgenesynteny
Dependencies
- Python 3.11
- requests
- packaging
- biopython
- configparser
- matplotlib
Getting started
- Install and open Miniconda
- Create an environment with python 3.11 e.g:
conda create -n ezgenesyntenyenv python=3.11
- Activate (enter) the environment:
conda activate ezgenesyntenyenv
- Install the package (this automatically installs all dependencies as well):
pip install ezgenesynteny
- Navigate to the folder you want to deposit the output files in:
cd \path\to\working\directory
- Then you're ready to begin!
ezgenesynteny usage
$ ezgenesynteny -h
usage: ezgenesynteny [-h] [-s SPECIES [SPECIES ...]] [-g GENE_NAME] -up UPGENES -down DOWNGENES
[-plot PLOTNAME] [-csv CSVNAME] [-f INPUT_FILE]
Query the GenBank database with species and gene names to obtain a list of genes upstream
and downstream of target gene.
options:
-h, --help show this help message and exit
-s SPECIES [SPECIES ...], --species SPECIES [SPECIES ...]
Species name(s) (e.g., 'Homo_sapiens' or 'Human')
-g GENE_NAME, --gene_name GENE_NAME
Gene name (e.g. BRCA1 or brca1)
-up UPGENES, --upgenes UPGENES
Number of upstream genes to search for
-down DOWNGENES, --downgenes DOWNGENES
Number of downstream genes to search for
-plot PLOTNAME, --plotname PLOTNAME
Output file name for the gene order plot
-csv CSVNAME, --csvname CSVNAME
Output file name for the gene order CSV
-f INPUT_FILE, --input_file INPUT_FILE
Path to a text file containing a list of species and genes
ezgenesynteny inputs:
-s, -g, -up, -down
The simple command line inputs are the species name (-s) and gene name (-g), and then the number of upstream (-up) and downstream (-down) genes to be obtained. For example, to get the human gdf5 gene with 5 upstream genes and downstream:
$ ezgenesynteny -s human -g gdf5 -up 5 -down 5
This produces the following output in the terminal:
▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒
▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒ ezgenesynteny: human gdf5 ▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒
▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒▒
Gene info:
Name: GDF5
Description: growth differentiation factor 5
Synonyms: ['OS5', 'LAP4', 'BDA1C', 'BMP14', 'CDMP1', 'LAP-4', 'SYM1B', 'SYNS2', 'BMP-14',
'DUPANS']
Locus: 20q11.22
Strand: reverse
Using record 0:
Organism: human (Homo sapiens)
Assembly: Chromosome 20 Reference GRCh38.p14 Primary Assembly
Accession: NC_000020
Location: 35433347:35454749
Length: 21403bp
Parsing upstream genomic record (35454750:40454749)...
Record file parsed in 2.8 seconds
Genes in region: {'CEP250': '+', 'C20orf173': '-', 'ERGIC3': '+', 'SPAG4': '+', 'CPNE1':
'-', 'RBM12': '-', 'NFS1': '-', 'ROMO1': '+', 'RBM39': '-', 'PHF20': '+', 'SCAND1': '-',
'CNBD2': '+', 'EPB41L1': '+', 'AAR2': '+', 'DLGAP4': '+', 'MYL9': '+', 'TGIF2': '+',
'TGIF2-RAB5IF': '+', 'RAB5IF': '+', 'SLA2': '-', 'NDRG3': '-', 'DSN1': '-', 'MTCL2': '-',
'TLDC2': '+', 'SAMHD1': '-', 'RBL1': '-', 'MROH8': '-', 'RPN2': '+', 'GHRH': '-',
'MANBAL': '+', 'SRC': '+', 'BLCAP': '-', 'NNAT': '+', 'CTNNBL1': '+', 'VSTM2L': '+',
'TTI1': '-', 'RPRD1B': '+', 'TGM2': '-', 'KIAA1755': '-', 'BPI': '+', 'LBP': '+',
'LOC124904958': '+', 'RALGAPB': '+', 'ADIG': '+', 'ARHGAP40': '+', 'SLC32A1': '+',
'ACTR5': '+', 'PPP1R16B': '+', 'FAM83D': '+', 'DHX35': '+'}
Parsing downstream genomic record (30433347:35433346)...
Record file parsed in 2.6 seconds
Genes in region: {'LOC124904970': '+', 'DEFB115': '+', 'DEFB116': '-', 'DEFB118': '+',
'DEFB119': '-', 'DEFB121': '-', 'DEFB123': '+', 'DEFB124': '-', 'REM1': '+', 'HM13': '+',
'MCTS2': '+', 'ID1': '+', 'COX4I2': '+', 'BCL2L1': '-', 'TPX2': '+', 'MYLK2': '+',
'FOXS1': '-', 'DUSP15': '-', 'TTLL9': '+', 'PDRG1': '-', 'XKR7': '+', 'CCM2L': '+',
'HCK': '+', 'TM9SF4': '+', 'PLAGL2': '-', 'POFUT1': '+', 'KIF3B': '+', 'ASXL1': '+',
'NOL4L': '-', 'C20orf203': '-', 'COMMD7': '-', 'DNMT3B': '+', 'MAPRE1': '+', 'EFCAB8':
'+', 'SUN5': '-', 'BPIFB2': '+', 'BPIFB6': '+', 'BPIFB3': '+', 'BPIFB4': '+', 'BPIFA2':
'+', 'BPIFA3': '+', 'BPIFA1': '+', 'BPIFB1': '+', 'CDK5RAP1': '-', 'SNTA1': '-',
'CBFA2T2': '+', 'NECAB3': '-', 'C20orf144': '+', 'ACTL10': '+', 'E2F1': '-', 'PXMP4':
'-', 'ZNF341': '+', 'CHMP4B': '+', 'RALY': '+', 'EIF2S2': '-', 'ASIP': '+', 'AHCY': '-',
'ITCH': '+', 'DYNLRB1': '+', 'MAP1LC3A': '+', 'PIGU': '-', 'TP53INP2': '+', 'NCOA6': '-',
'GGT7': '-', 'ACSS2': '+', 'GSS': '-', 'MYH7B': '+', 'TRPC4AP': '-', 'MMP24-AS1-EDEM2':
'-', 'EDEM2': '-', 'PROCR': '+', 'MMP24': '+', 'MMP24OS': '-', 'EIF6': '-', 'FAM83C': '-',
'UQCC1': '-'}
Trimming to max. 5 upstream genes and max. 5 downstream genes with GDF5 on forward strand:
[('CPNE1', '+'), ('SPAG4', '-'), ('ERGIC3', '-'), ('C20orf173', '+'), ('CEP250', '-'),
('GDF5', '+'), ('UQCC1', '+'), ('FAM83C', '+'), ('EIF6', '+'), ('MMP24OS', '+'), ('MMP24',
'-')]
Completed in 19.3 seconds
The species name can be entered either as the common name (human) or latin name (homo_sapiens). In latin names or common names with multiple words, underscores must be used to separate the words (e.g. carcharodon_carcharias or great_white_shark). A wide variety of gene synonyms can be searched for rather than the specific or canonical gene symbol. It's important to be aware of this - for example, if you search for the zebrafish shh gene. In this case, as a result of the teleost whole-genome duplication, zebrafish possess two paralogs of shh: shha and shhb, but the result will be the shha gene only, because shh is listed as one of its synonyms while it isn't for shhb.
You can also search for genes using refseq transcript or protein IDs (e.g. NM_008109.4 or NP_032135.2), but be aware that the species associated with these IDs will take precedence over the species given as an argument in -s. For example, NM_008109.4 is from the mouse genome, so even if the command given is -s human -g NM_008109.4, the result will be the mouse gene, so be careful to check the "Organism" line that gets printed in the terminal to be sure. The way the GenBank search works is essentially the same as writing e.g. "human gdf5" into the search bar on the NCBI gene website, so you can test a query there if you encounter a problem.
Details about the target gene are printed in the terminal, followed by lists of upstream and downstream protein-coding genes (from the perspective of the target gene's orentiation). The searched regions in these case are 5Mb upstream and downstream, because the region lengths are specified by 1Mb multiplied by the number of genes specified by -up and -down (in this example 5). Finally, these long lists of genes are trimmed to the specified number, producing in this example an ordered list of 11 genes representing the 5 upstream genes, the target gene, and the 5 downstream genes, including the strand orientation of each gene.
You can enter multiple species names to get an easy comparison of the local gene synteny around homologous target genes. For example:
$ ezgenesynteny -s human mouse chicken -g gdf5 -up 5 -down 5
The program loops through the different species and prints each output as above to the terminal.
-f
Alternatively, a set of species and gene names can be input from a plain text file in the working directory filepath, where the text file looks like this:
human, nkx2-5
mouse, nkx2-5
stegostoma_tigrinum, LOC125458387
This input method is particularly useful for cases like the one above where the homologous target gene has significantly different names in different species. Searching for the nkx2-5 gene in human and mouse returns the gene, but returns no results in the zebra shark (stegostoma_tigrinum), because it's not completely annotated in the genome assembly of this species. When running the program with an input text file like this, it loops through each species with its corresponding gene name and prints the results to the terminal e.g:
$ ezgenesynteny -f inputfile.txt -up 5 -down 5
-plot
To visualise the gene order/synteny data as a gene arrow map (saved to an image file), add the -plot command with the desired output file name with format suffix, for example:
$ ezgenesynteny -f inputfile.txt -up 5 -down 5 -plot nkx2-5plot.png
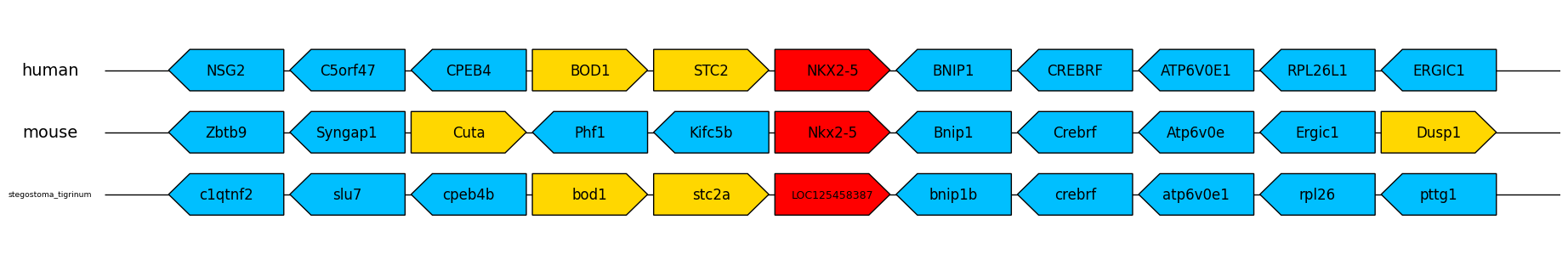
When this command is run, the program prints the results to the terminal as usual, but also saves this image to the working directory:
The target gene is coloured in red, and then forward/reverse strand genes are coloured in yellow/blue. A wide range of image formats are supported, including PDF and EPS which can be imported and edited in a vector graphics editor such as Adobe Illustrator or GIMP. If no format suffix is specified (e.g. '-plot nkx2-5plot'), PDF is selected as default.
-csv
To save the gene order/synteny data to a CSV file, add the -csv command with the desired CSV filename (without suffix), for example:
$ ezgenesynteny -f inputfile.txt -up 5 -down 5 -plot nkx2-5plot.png -csv nkx2-5csv
This can be particularly useful for manually editing the spacing of the genes in a table to align the homologous genes.
changeemail usage
$ changeemail -h
usage: changeemail [-h] [-check] [-update]
Manage email address for GenBank Entrez queries. Only stored locally and sent with
queries to NCBI, nowhere else.
options:
-h, --help show this help message and exit
-check Check the current saved email address
-update Update the email address
The first time you try to run ezgenesyteny, you will be prompted for an email address as NCBI requires this to send queries via Entrez. After you enter this email address the first time, you won't need to enter it again, as it saved locally to a config file in the package directory. It is never shared with anyone except NCBI when you send queries via Entrez. This accessory module provides the option to check or update (change) this email address if you decide to use a different address at a later date. Any dummy email address or word (e.g. 'dummy@gmail.com' or 'dummy') can also be used if preferred.
Bugs
Please submit via the GitHub issues page.
Software Licence
Project details
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
Hashes for ezgenesynteny-0.0.1-py3-none-any.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 0662d5394b3d527306ebc4ff37ee8e1eb071d3564ef0e63be4b8a5e885dcae02 |
|
| MD5 | cd31fc493523e07a7358142cc2c5131e |
|
| BLAKE2b-256 | 1a344541a80cd0b7ebed2f94f49154f56bc38dbf5dc16e6a00e76e298d4cc530 |