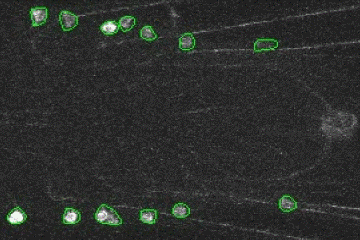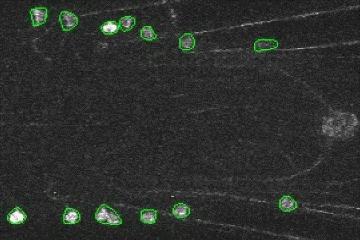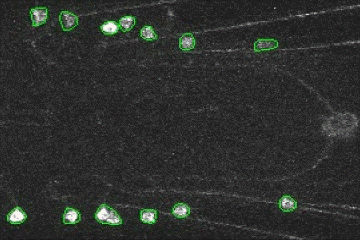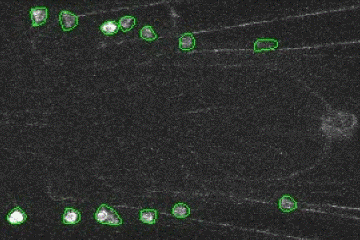
Fluorescence Signal Analyzer for calcium imaging on user-defined neural structures.
Project description
FluoSA (Fluorescence Signal Analyzer)
FluoSA inputs *.LIF and *.tif files, detects user-defined neural structures, and quantifies the fluorescence signal changes (frame-wise fluorescence intensity and the dF/F0) in these structures.
The outputs of FluoSA include:
-
An annotated video showing the detected neural structures:
-
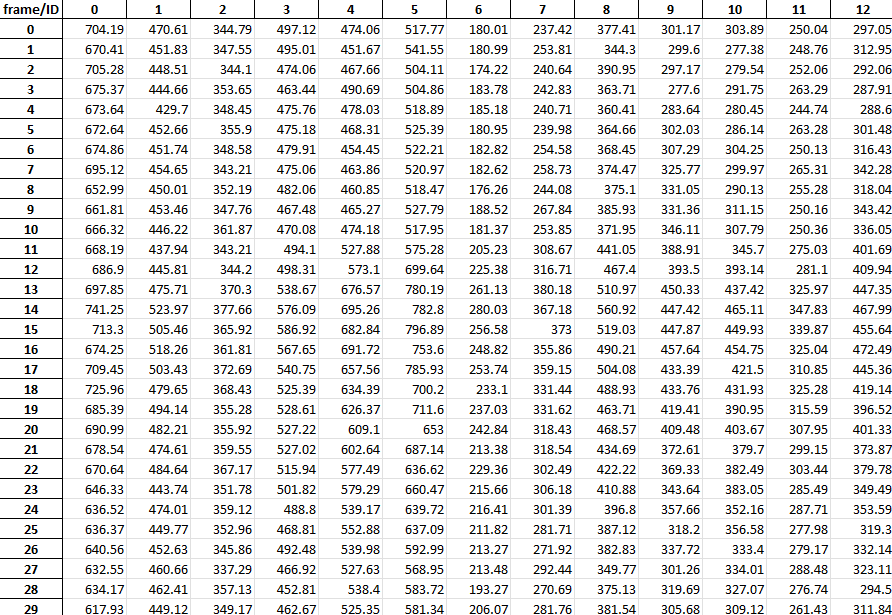
Spreadsheets storing frame-wise fluorescence intensity in each detected neural structure:
-
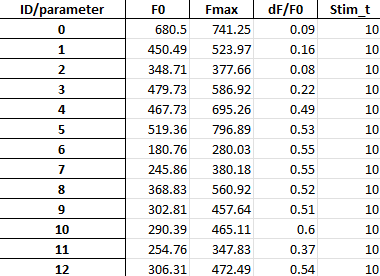
Spreadsheets storing summary of fluorescence signal changes (dF/F0) in each detected neural structure:
Installation
1 Install FluoSA
python3 -m pip install FluoSA
2 Install CUDA toolkit v11.8
https://developer.nvidia.com/cuda-11-8-0-download-archive?target_os=Windows&target_arch=x86_64
3 Install Detectron2
python3 -m pip install 'git+https://github.com/facebookresearch/detectron2.git'
4 Install PyTorch 2.0.1
4.1 For Windows and Linux
4.1.1 If using GPU
python3 -m pip install torch==2.0.1 torchvision==0.15.2 torchaudio==2.0.2 --index-url https://download.pytorch.org/whl/cu118
4.1.2 CPU only
python3 -m pip install torch==2.0.1 torchvision==0.15.2 torchaudio==2.0.2 --index-url https://download.pytorch.org/whl/cpu
4.2 For Mac
python3 -m pip install torch==2.0.1 torchvision==0.15.2 torchaudio==2.0.2
Usage
In your terminal / command prompt, type:
FluoSA
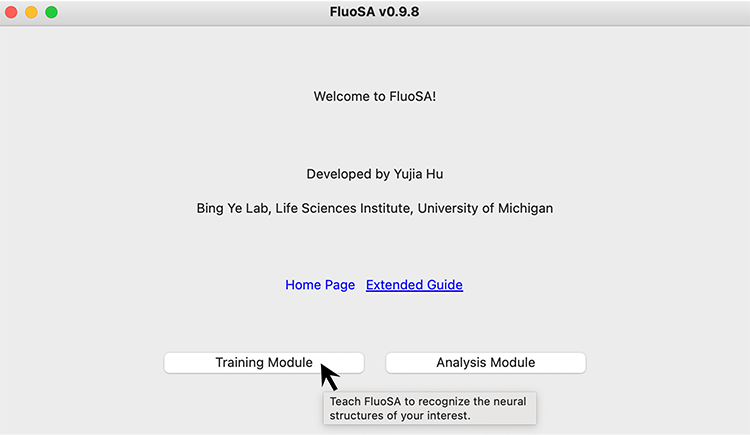
Then the user interface will be initiated:
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
Filter files by name, interpreter, ABI, and platform.
If you're not sure about the file name format, learn more about wheel file names.
Copy a direct link to the current filters
File details
Details for the file fluosa-1.0.0.tar.gz.
File metadata
- Download URL: fluosa-1.0.0.tar.gz
- Upload date:
- Size: 26.9 kB
- Tags: Source
- Uploaded using Trusted Publishing? Yes
- Uploaded via: twine/5.1.1 CPython/3.12.5
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
a7abe905e9fa8a2ff1c5454a3aa18307bde7d207393ff733854d7d1469586e0b
|
|
| MD5 |
2418715a2f69e364465078904a801e97
|
|
| BLAKE2b-256 |
fff3c07ae8e5adde804c0e1f367e62565d892bdea820d077dc149f6fb7a9a568
|
File details
Details for the file fluosa-1.0.0-py3-none-any.whl.
File metadata
- Download URL: fluosa-1.0.0-py3-none-any.whl
- Upload date:
- Size: 31.6 kB
- Tags: Python 3
- Uploaded using Trusted Publishing? Yes
- Uploaded via: twine/5.1.1 CPython/3.12.5
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
0ba44f951a6df4e47859ccf03ab9628bf461b08828bf750562d48b74fdddf3bb
|
|
| MD5 |
4967c0e39c71b752c6c5c55ce12f70c1
|
|
| BLAKE2b-256 |
0943a152f7be138ad923e1d649c141b59db240989bcc79de6dcde53ebea91903
|