A quick and precise pipeline for detecting phages in sequence assemblies.
Project description
.
,'/ \`.
|\/___\/|
\'\ /`/ ██╗ █████╗ ███████╗ ██████╗ ███████╗██████╗
`.\ /,' ██║██╔══██╗██╔════╝██╔════╝ ██╔════╝██╔══██╗
| ██║███████║█████╗ ██║ ███╗█████╗ ██████╔╝
| ██ ██║██╔══██║██╔══╝ ██║ ██║██╔══╝ ██╔══██╗
|=| ╚█████╔╝██║ ██║███████╗╚██████╔╝███████╗██║ ██║
/\ ,|=|. /\ ╚════╝ ╚═╝ ╚═╝╚══════╝ ╚═════╝ ╚══════╝╚═╝ ╚═╝
,'`. \/ |=| \/ ,'`.
,' `.|\ `-' /|,' `.
,' .-._ \ `---' / _,-. `.
,' `-`-._,-'-' `.
'
Jaeger : A quick and precise pipeline for detecting phages in sequence assemblies.
Jaeger is a tool that utilizes homology-free machine learning to identify phage genome sequences that are hidden within metagenomes. It is capable of detecting both phages and prophages within metagenomic assemblies.
visit our wiki: https://github.com/Yasas1994/Jaeger/wiki/%22Jaeger:-yet-AnothEr-phaGE-pRedictor%22
Installation
Seting up the environment (Linux)
Jaeger is currently tested only on python 3.9.2 therefore, we recomend you to setup a conda environmet by running
for CPU only version
conda create -n jaeger python=3.9
conda install -n jaeger 'tensorflow>=2.10' 'numpy>=1.19.5' 'tqdm>=4.64.0' biopython=1.78
for GPU version
The performance of the Jaeger workflow can be significantly increased by utilizing GPUs. By adding more GPUs to the system, the speedup can be linear. To enable GPU support, the CUDA Toolkit and cuDNN library must be installed in the created conda environment. (Note: This step can be skipped if you do not wish to use a GPU.) It is important to ensure that the versions of CUDA Toolkit and cuDNN are compatible with the version of CUDA installed on your system.
if you have cuda=10.1 installed on your system,
conda create -n jaeger python=3.9 pip
conda install -n jaeger -c conda-forge cudatoolkit=10.1
conda install -n jaeger -c conda-forge cudnn=7.6.5.32
conda install -n jaeger 'tensorflow>=2.10' 'numpy>=1.19.5' 'tqdm>=4.64.0' biopython=1.78
if you have cuda>=11.1 installed on your system,
conda create -n jaeger python=3.9 pip
conda install -n jaeger -c "nvidia/label/cuda-11.x.x" cudatoolkit=11
conda install -n jaeger -c conda-forge cudnn
conda activate jaeger #or source activate path/to/jaeger/env depending on your cluster configuration
pip install 'tensorflow>=2.10' 'numpy>=1.19.5' 'tqdm>=4.64' 'biopython>=1.78'
Setting up the environment (Apple silicon)
conda create -c conda-forge -c apple -c bioconda -c defaults -n jaeger python=3.9.2 pip tensorflow=2.6 tensorflow-deps=2.6.0 numpy=1.19.5 tqdm=4.64.0 biopython=1.78
to add support for apple silicon gpus run
conda activate jaeger
pip install tensorflow-macos
pip install tensorflow-metal
finally, clone the repository by running
git clone https://github.com/Yasas1994/Jaeger
and install Jaeger
pip install ./Jaeger/.
Environmnet variables
- Tensorflow logging behaviour can be altered by 'TF_CPP_MIN_LOG_LEVEL' environment variable. Jaeger's default log level is 3
export TF_CPP_MIN_LOG_LEVEL=3
| levels | description |
|---|---|
| 0 | all messages are logged (default behavior) |
| 1 | INFO messages are not printed |
| 2 | INFO and WARNING messages are not printed |
| 3 | INFO, WARNING, and ERROR messages are not printed |
Running Jaeger
Once the environment is properly set up, using Jaeger is straightforward. The program can accept both compressed and uncompressed FASTA files containing the contigs as input. It will output a table containing the predictions and various statistics calculated during runtime. By default, Jaeger will run on all GPUs present on the system, however, this behavior can be overridden by providing a list of GPU names to the -gnames option, which limits the number of GPUs available for Jaeger's use. The -ofasta option will write all potential viral contigs (contigs that meet the cutoff score) into a separate file.
Jaeger -i input_file.fasta -o output_dir --batch 128
NEW : Running Jaeger in multi-GPU mode
We provide a new program that allows users to automatically run multiple instances of Jaeger on several GPUs allowing maximum utilization of state-of-the-art hardware. This program accepts a csv file with paths to all input fasta files. Column with the file paths should be named as 'paths'. All other arguments reamains similar to 'Jaeger' program.
Jaeger_parallel -i input_file.csv -o output_dir --batch 128
Selecting batch parameter
You can control the number of parallel computations using this parameter. By default it is set to 512. If you run into OOM errors, please consider setting the --bactch option to a lower value. for example 128 is good enough for a grraphics card with 6 Gb of memory.
options
options:
-h, --help show this help message and exit
-i INPUT, --input INPUT
path to input file
-o OUTPUT, --output OUTPUT
path to output directory
-of OFASTA, --ofasta OFASTA
path to output fasta file
--cutoff CUTOFF fasta output cutoff score
--fsize [FSIZE] length of the sliding window (value must be 2^n). default:2048
--stride [STRIDE] stride of the sliding window. default:2048 (stride==fsize)
-m {default,experimental_1,experimental_2}, --model {default,experimental_1,experimental_2}
select a deep-learning model to use. default:default
--batch [BATCH] parallel batch size, set to a lower value if your gpu runs out of memory. default:128
--getalllogits return position-wise logits for each prediction window as a .npy file
--usecutoffs use cutoffs to obtain the class prediction
--cpu ignore available gpus and explicitly run jaeger on cpu. default: False
--virtualgpu create and run jaeger on a virtualgpu. default: False
--physicalid [PHYSICALID]
sets the default gpu device id (for multi-gpu systems). default:0
--getalllabels get predicted labels for Non-Viral contigs. default:False
--meanscore output mean predictive score per contig. deafault:True
--fragscore output percentage of perclass predictions per contig. deafault:True
Misc. Options:
-v, --verbose Verbosity level : -v warning, -vv info, -vvv debug, (default info)
-f, --overwrite Overwrite existing files
--progressbar show progress bar
Notes
- The program expects the input file to be in FASTA format.
- The program uses a sliding window approach to scan the input sequences, so the stride argument determines how far the window will move after each scan.
- The batch argument determines how many sequences will be processed in parallel.
- The program is compatible with both CPU and GPU. By default, it will run on the GPU, but if the --cpu option is provided, it will use the specified number of threads for inference.
- The program uses a pre-trained neural network model for phage genome prediction.
- The --getalllabels option will output predicted labels for Non-Viral contigs, which can be useful for further analysis. It's recommended to use the output of this program in conjunction with other methods for phage genome identification.
Comming soon - Python Library
Jaeger can be integreted into python scripts using the jaegeraa python library as follows. currenly the predict function accepts 4 diffent input types.
- Nucleotide sequence -> str
- List of Nucleotide sequences -> list(str,str,..)
- python fileobject -> (io.TextIOWrapper)
- python generator object that yields Nucleotide sequences as str (types.GeneratorType)
- Biopython Seq object
from jaegeraa.api import Predictions
model=Predictor()
predictions=model.predict(input,stride=2048,fragsize=2048,batch=100)
model.predict() returns a dictionary of lists in the following format
{'contig_id': ['seq_0', 'seq_1'],
'length': [19000, 10503],
'#num_prok_windows': [0, 0],
'#num_vir_windows': [9, 0],
'#num_fun_windows': [0, 5],
'#num_arch_windows': [0, 0],
'prediction': ['Phage', 'Non-phage'],
'bac_score': [-1.9552012549506292, -1.9441368103027343],
'vir_score': [6.6312947273254395, -3.097817325592041],
'fun_score': [-5.712721400790745, -0.6870137214660644],
'arch_score': [-2.4369852013058133, -0.8941479325294495],
'window_summary': ['9V', '5n']}
This dictionary can be easily converted to a pandas dataframe using DataFrame.from_dict() method
import pandas as pd
df = DataFrame.from_dict(predictions)
What is in the output?
| contig_id | length | #num_prok_windows | #num_vir_windows | #num_fun_windows | #num_arch_windows | prediction | bac_score | vir_score | fun_score | arch_score | window_summary |
|---|---|---|---|---|---|---|---|---|---|---|---|
| NODE_21_length_108942_cov_81.621865 | 108942 | 0 | 53 | 0 | 0 | Phage | -0.2883 | 5.1653 | -6.4840 | -4.5684 | 53V |
| NODE_85_length_39232_cov_116.902519 | 39232 | 0 | 19 | 0 | 0 | Phage | 0.3820 | 4.0256 | -5.5622 | -3.6816 | 19V |
| NODE_151_length_25306_cov_102.578472 | 25306 | 0 | 12 | 0 | 0 | Phage | -0.0779 | 5.0276 | -6.1431 | -4.0177 | 12V |
| NODE_214_length_19298_cov_103.990178 | 19298 | 0 | 9 | 0 | 0 | Phage | 0.3472 | 4.9460 | -6.3656 | -4.5528 | 9V |
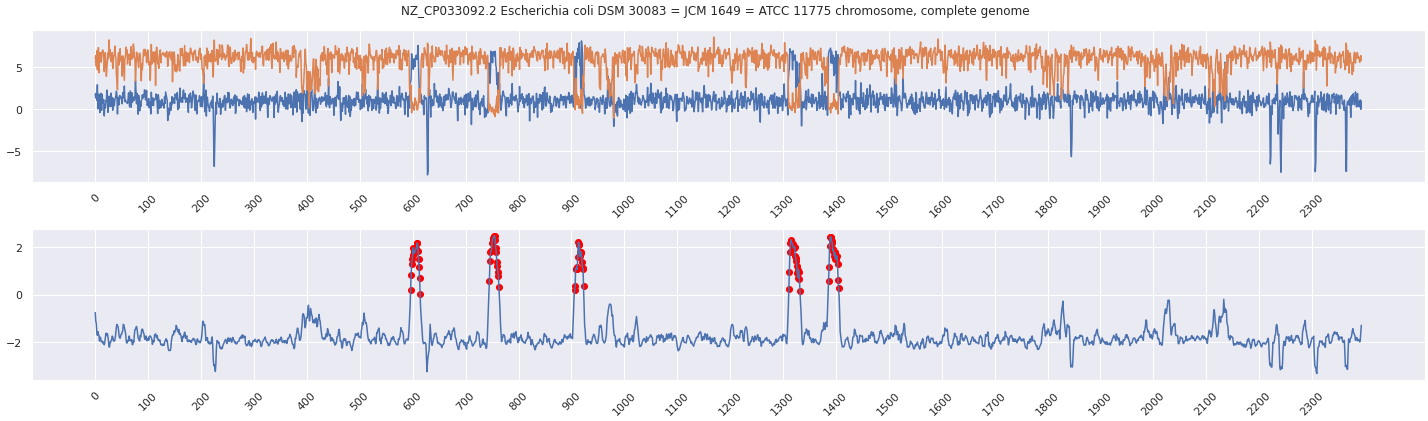
This table provides information about various contigs in a metagenomic assembly. Each row represents a single contig, and the columns provide information about the contig's ID, length, the number of windows identified as prokaryotic, viral, eukaryotic, and archaeal, the prediction of the contig (Phage or Non-phage), the score of the contig for each category (bacterial, viral, eukaryotic and archaeal), and a summary of the windows. The table can be used to identify potential phage sequences in the metagenomic assembly based on the prediction column. The score columns can be used to further evaluate the confidence of the prediction and the window summary column can be used to understand the count of windows that contributed to the final prediction.
Comming soon - Predicting prophages with Jaeger
Visualizing predictions
You can use phage_contig_annotator to annotate and visualize Jaeger predictions.
Always keep in mind that ...
Acknowlegements
This work was supported by the European Union’s Horizon 2020 research and innovation program, under the Marie Skłodowska-Curie Actions Innovative Training Networks grant agreement no. 955974 (VIROINF), the European Research Council (ERC) Consolidator grant 865694
ascii art from https://ascii.co.uk/
Project details
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distributions
Built Distribution
File details
Details for the file jaeger_bio-1.1.24-py3-none-any.whl.
File metadata
- Download URL: jaeger_bio-1.1.24-py3-none-any.whl
- Upload date:
- Size: 33.1 MB
- Tags: Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/5.0.0 CPython/3.9.18
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 1c37e2185fa5d439d4d170f0582db6aee4bd1c5f8ed9285663aff867c7049497 |
|
| MD5 | f5c3991688da053933bffdb9522f11e2 |
|
| BLAKE2b-256 | fc1de19229335e7fcef34753dcc33873ff17c0aeccbeb44116d68f217c491895 |