Test
Project description
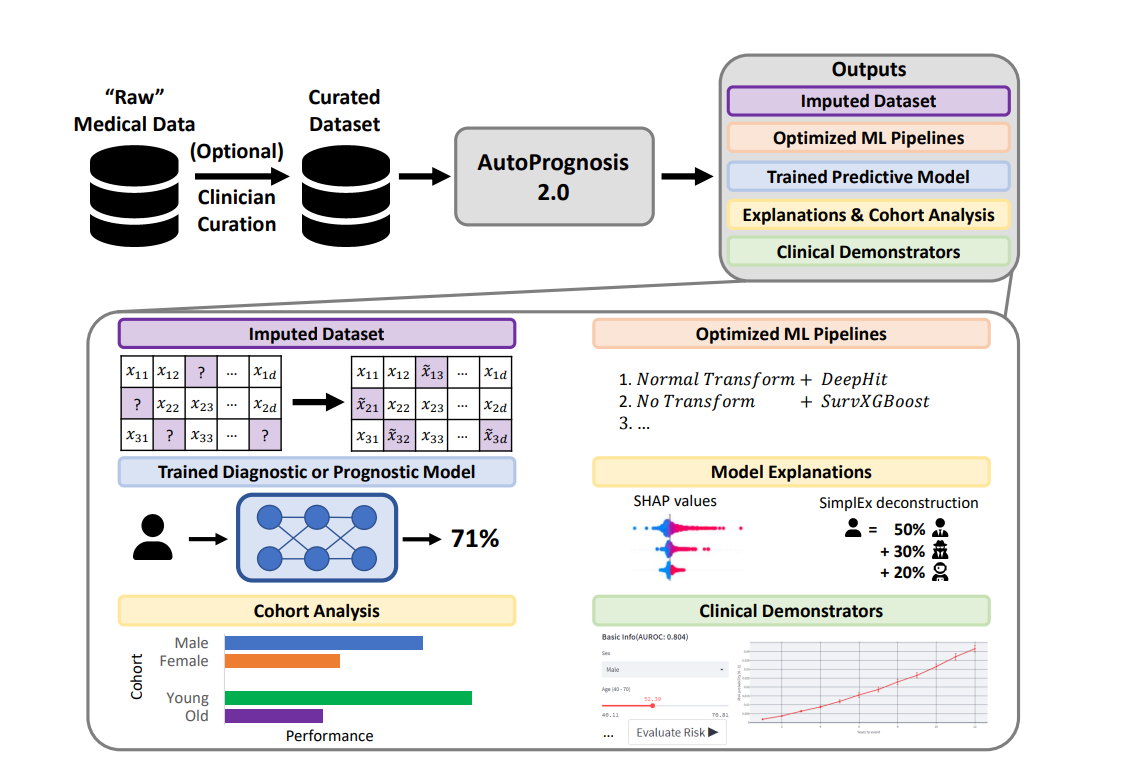
Jori AutoPrognosis (FORKED) - A system for automating the design of predictive modeling pipelines tailored for clinical prognosis.
:key: Features
- :rocket: Automatically learns ensembles of pipelines for classification, regression or survival analysis tasks.
- :cyclone: Easy to extend pluginable architecture.
- :fire: Interpretability and uncertainty quantification tools.
- :adhesive_bandage: Data imputation using HyperImpute.
- :zap: Build demonstrators using Streamlit.
- :notebook: Python and R tutorials available.
- :book: Read the docs
Note : The R bindings have been tested using R version 4.2 and Python 3.8.
:rocket: Installation
Using pip
The library can be installed from PyPI using
$ pip install autoprognosis
or from source, using
$ pip install .
Redis (Optional, but recommended)
AutoPrognosis can use Redis as a backend to improve the performance and quality of the searches.
For that, install the redis-server package following the steps described on the official site.
Environment variables
The library can be configured from a set of environment variables.
| Variable | Description |
|---|---|
N_OPT_JOBS |
Number of cores to use for hyperparameter search. Default : 1 |
N_LEARNER_JOBS |
Number of cores to use by inidividual learners. Default: all cpus |
REDIS_HOST |
IP address for the Redis database. Default 127.0.0.1 |
REDIS_PORT |
Redis port. Default: 6379 |
Example: export N_OPT_JOBS = 2 to use 2 cores for hyperparam search.
:boom: Sample Usage
Advanced Python tutorials can be found in the Python tutorials section.
R examples can be found in the R tutorials section.
List the available classifiers
from autoprognosis.plugins.prediction.classifiers import Classifiers
print(Classifiers().list_available())
Create a study for classifiers
from sklearn.datasets import load_breast_cancer
from autoprognosis.studies.classifiers import ClassifierStudy
from autoprognosis.utils.serialization import load_model_from_file
from autoprognosis.utils.tester import evaluate_estimator
X, Y = load_breast_cancer(return_X_y=True, as_frame=True)
df = X.copy()
df["target"] = Y
study_name = "example"
study = ClassifierStudy(
study_name=study_name,
dataset=df, # pandas DataFrame
target="target", # the label column in the dataset
)
model = study.fit()
# Predict the probabilities of each class using the model
model.predict_proba(X)
(Advanced) Customize the study for classifiers
from pathlib import Path
from sklearn.datasets import load_breast_cancer
from autoprognosis.studies.classifiers import ClassifierStudy
from autoprognosis.utils.serialization import load_model_from_file
from autoprognosis.utils.tester import evaluate_estimator
X, Y = load_breast_cancer(return_X_y=True, as_frame=True)
df = X.copy()
df["target"] = Y
workspace = Path("workspace")
study_name = "example"
study = ClassifierStudy(
study_name=study_name,
dataset=df, # pandas DataFrame
target="target", # the label column in the dataset
num_iter=100, # how many trials to do for each candidate
timeout=60, # seconds
classifiers=["logistic_regression", "lda", "qda"],
workspace=workspace,
)
study.run()
output = workspace / study_name / "model.p"
model = load_model_from_file(output)
# <model> contains the optimal architecture, but the model is not trained yet. You need to call fit() to use it.
# This way, we can further benchmark the selected model on the training set.
metrics = evaluate_estimator(model, X, Y)
print(f"model {model.name()} -> {metrics['str']}")
# Train the model
model.fit(X, Y)
# Predict the probabilities of each class using the model
model.predict_proba(X)
List the available regressors
from autoprognosis.plugins.prediction.regression import Regression
print(Regression().list_available())
Create a Regression study
# third party
import pandas as pd
# autoprognosis absolute
from autoprognosis.utils.serialization import load_model_from_file
from autoprognosis.utils.tester import evaluate_regression
from autoprognosis.studies.regression import RegressionStudy
# Load dataset
df = pd.read_csv(
"https://archive.ics.uci.edu/ml/machine-learning-databases/00291/airfoil_self_noise.dat",
header=None,
sep="\\t",
)
last_col = df.columns[-1]
y = df[last_col]
X = df.drop(columns=[last_col])
df = X.copy()
df["target"] = y
# Search the model
study_name="regression_example"
study = RegressionStudy(
study_name=study_name,
dataset=df, # pandas DataFrame
target="target", # the label column in the dataset
)
model = study.fit()
# Predict using the model
model.predict(X)
Advanced Customize the Regression study
# stdlib
from pathlib import Path
# third party
import pandas as pd
# autoprognosis absolute
from autoprognosis.utils.serialization import load_model_from_file
from autoprognosis.utils.tester import evaluate_regression
from autoprognosis.studies.regression import RegressionStudy
# Load dataset
df = pd.read_csv(
"https://archive.ics.uci.edu/ml/machine-learning-databases/00291/airfoil_self_noise.dat",
header=None,
sep="\\t",
)
last_col = df.columns[-1]
y = df[last_col]
X = df.drop(columns=[last_col])
df = X.copy()
df["target"] = y
# Search the model
workspace = Path("workspace")
workspace.mkdir(parents=True, exist_ok=True)
study_name="regression_example"
study = RegressionStudy(
study_name=study_name,
dataset=df, # pandas DataFrame
target="target", # the label column in the dataset
num_iter=10, # how many trials to do for each candidate. Default: 50
num_study_iter=2, # how many outer iterations to do. Default: 5
timeout=50, # timeout for optimization for each classfier. Default: 600 seconds
regressors=["linear_regression", "xgboost_regressor"],
workspace=workspace,
)
study.run()
# Test the model
output = workspace / study_name / "model.p"
model = load_model_from_file(output)
# <model> contains the optimal architecture, but the model is not trained yet. You need to call fit() to use it.
# This way, we can further benchmark the selected model on the training set.
metrics = evaluate_regression(model, X, y)
print(f"Model {model.name()} score: {metrics['str']}")
# Train the model
model.fit(X, y)
# Predict using the model
model.predict(X)
List available survival analysis estimators
from autoprognosis.plugins.prediction.risk_estimation import RiskEstimation
print(RiskEstimation().list_available())
Create a Survival analysis study
# third party
import numpy as np
from pycox import datasets
# autoprognosis absolute
from autoprognosis.studies.risk_estimation import RiskEstimationStudy
from autoprognosis.utils.serialization import load_model_from_file
from autoprognosis.utils.tester import evaluate_survival_estimator
df = datasets.gbsg.read_df()
df = df[df["duration"] > 0]
X = df.drop(columns = ["duration"])
T = df["duration"]
Y = df["event"]
eval_time_horizons = np.linspace(T.min(), T.max(), 5)[1:-1]
study_name = "example_risks"
study = RiskEstimationStudy(
study_name=study_name,
dataset=df,
target="event",
time_to_event="duration",
time_horizons=eval_time_horizons,
)
model = study.fit()
# Predict using the model
model.predict(X, eval_time_horizons)
Advanced Customize the Survival analysis study
# stdlib
import os
from pathlib import Path
# third party
import numpy as np
from pycox import datasets
# autoprognosis absolute
from autoprognosis.studies.risk_estimation import RiskEstimationStudy
from autoprognosis.utils.serialization import load_model_from_file
from autoprognosis.utils.tester import evaluate_survival_estimator
df = datasets.gbsg.read_df()
df = df[df["duration"] > 0]
X = df.drop(columns = ["duration"])
T = df["duration"]
Y = df["event"]
eval_time_horizons = np.linspace(T.min(), T.max(), 5)[1:-1]
workspace = Path("workspace")
study_name = "example_risks"
study = RiskEstimationStudy(
study_name=study_name,
dataset=df,
target="event",
time_to_event="duration",
time_horizons=eval_time_horizons,
num_iter=10,
num_study_iter=1,
timeout=10,
risk_estimators=["cox_ph", "survival_xgboost"],
score_threshold=0.5,
workspace=workspace,
)
study.run()
output = workspace / study_name / "model.p"
model = load_model_from_file(output)
# <model> contains the optimal architecture, but the model is not trained yet. You need to call fit() to use it.
# This way, we can further benchmark the selected model on the training set.
metrics = evaluate_survival_estimator(model, X, T, Y, eval_time_horizons)
print(f"Model {model.name()} score: {metrics['str']}")
# Train the model
model.fit(X, T, Y)
# Predict using the model
model.predict(X, eval_time_horizons)
:high_brightness: Tutorials
Plugins
AutoML
Classification tasks
Classification tasks with imputation
Survival analysis tasks
Survival analysis tasks with imputation
Regression tasks
Classifiers with explainers
Multiple imputation example
Building a demonstrator
:zap: Plugins
Imputation methods
from autoprognosis.plugins.imputers import Imputers
imputer = Imputers().get(<NAME>)
| Name | Description |
|---|---|
| hyperimpute | Iterative imputer using both regression and classification methods based on linear models, trees, XGBoost, CatBoost and neural nets |
| mean | Replace the missing values using the mean along each column with SimpleImputer |
| median | Replace the missing values using the median along each column with SimpleImputer |
| most_frequent | Replace the missing values using the most frequent value along each column with SimpleImputer |
| missforest | Iterative imputation method based on Random Forests using IterativeImputer and ExtraTreesRegressor |
| ice | Iterative imputation method based on regularized linear regression using IterativeImputer and BayesianRidge |
| mice | Multiple imputations based on ICE using IterativeImputer and BayesianRidge |
| softimpute | Low-rank matrix approximation via nuclear-norm regularization |
| EM | Iterative procedure which uses other variables to impute a value (Expectation), then checks whether that is the value most likely (Maximization) - EM imputation algorithm |
| gain | GAIN: Missing Data Imputation using Generative Adversarial Nets |
Preprocessing methods
from autoprognosis.plugins.preprocessors import Preprocessors
preprocessor = Preprocessors().get(<NAME>)
| Name | Description |
|---|---|
| maxabs_scaler | Scale each feature by its maximum absolute value. MaxAbsScaler |
| scaler | Standardize features by removing the mean and scaling to unit variance. - StandardScaler |
| feature_normalizer | Normalize samples individually to unit norm. Normalizer |
| normal_transform | Transform features using quantiles information.QuantileTransformer |
| uniform_transform | Transform features using quantiles information.QuantileTransformer |
| minmax_scaler | Transform features by scaling each feature to a given range.MinMaxScaler |
Classification
from autoprognosis.plugins.prediction.classifiers import Classifiers
classifier = Classifiers().get(<NAME>)
| Name | Description |
|---|---|
| neural_nets | PyTorch based neural net classifier. |
| logistic_regression | LogisticRegression |
| catboost | Gradient boosting on decision trees - CatBoost |
| random_forest | A random forest classifier. RandomForestClassifier |
| tabnet | TabNet : Attentive Interpretable Tabular Learning |
| xgboost | XGBoostClassifier |
Survival Analysis
from autoprognosis.plugins.prediction.risk_estimation import RiskEstimation
predictor = RiskEstimation().get(<NAME>)
| Name | Description |
|---|---|
| survival_xgboost | XGBoost Survival Embeddings |
| loglogistic_aft | Log-Logistic AFT model |
| deephit | DeepHit: A Deep Learning Approach to Survival Analysis with Competing Risks |
| cox_ph | Cox’s proportional hazard model |
| weibull_aft | Weibull AFT model. |
| lognormal_aft | Log-Normal AFT model |
| coxnet | CoxNet is a Cox proportional hazards model also referred to as DeepSurv |
Regression
from autoprognosis.plugins.prediction.regression import Regression
regressor = Regression().get(<NAME>)
| Name | Description |
|---|---|
| tabnet_regressor | TabNet : Attentive Interpretable Tabular Learning |
| catboost_regressor | Gradient boosting on decision trees - CatBoost |
| random_forest_regressor | RandomForestRegressor |
| xgboost_regressor | XGBoostClassifier |
| neural_nets_regression | PyTorch-based neural net regressor. |
| linear_regression | LinearRegression |
Explainers
from autoprognosis.plugins.explainers import Explainers
explainer = Explainers().get(<NAME>)
| Name | Description |
|---|---|
| risk_effect_size | Feature importance using Cohen's distance between probabilities |
| lime | Lime: Explaining the predictions of any machine learning classifier |
| symbolic_pursuit | [Symbolic Pursuit](Learning outside the black-box: at the pursuit of interpretable models) |
| shap_permutation_sampler | SHAP Permutation Sampler |
| kernel_shap | SHAP KernelExplainer |
| invase | INVASE: Instance-wise Variable Selection |
Uncertainty
from autoprognosis.plugins.uncertainty import UncertaintyQuantification
model = UncertaintyQuantification().get(<NAME>)
| Name | Description |
|---|---|
| cohort_explainer | |
| conformal_prediction | |
| jackknife |
:hammer: Test
After installing the library, the tests can be executed using pytest
$ pip install .[testing]
$ pytest -vxs -m "not slow"
Citing
If you use this code, please cite the associated paper:
@misc{https://doi.org/10.48550/arxiv.2210.12090,
doi = {10.48550/ARXIV.2210.12090},
url = {https://arxiv.org/abs/2210.12090},
author = {Imrie, Fergus and Cebere, Bogdan and McKinney, Eoin F. and van der Schaar, Mihaela},
keywords = {Machine Learning (cs.LG), Artificial Intelligence (cs.AI), FOS: Computer and information sciences, FOS: Computer and information sciences},
title = {AutoPrognosis 2.0: Democratizing Diagnostic and Prognostic Modeling in Healthcare with Automated Machine Learning},
publisher = {arXiv},
year = {2022},
copyright = {Creative Commons Attribution 4.0 International}
}
References
- AutoPrognosis: Automated Clinical Prognostic Modeling via Bayesian Optimization with Structured Kernel Learning
- Prognostication and Risk Factors for Cystic Fibrosis via Automated Machine Learning
- Cardiovascular Disease Risk Prediction using Automated Machine Learning: A Prospective Study of 423,604 UK Biobank Participants
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distributions
Built Distributions
Hashes for jori_autoprognosis-0.2.3-py2.py3-none-macosx_10_14_x86_64.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 5e474d47044908b85fc64505bc849c82e812194f52ed4b073a177181fffae567 |
|
| MD5 | fd52abaab78edcd2861fa0411adab42a |
|
| BLAKE2b-256 | e3de279ef66b79e9cd565d63af1aae96760ad62409a7e6c97860f14978495e81 |
Hashes for jori_autoprognosis-0.2.3-py2.py3-none-any.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | d2d915d7bf75e377280ee454fd03a4303cef631b0bc81d159e1181a3f1230763 |
|
| MD5 | e0b1f7a0d366e1c4aa3b3e96429eac13 |
|
| BLAKE2b-256 | c129ba8a3ebdd9f5707b52938dfd08d15c11fea704519dc0da1dd4ad0db11dd2 |