Creating figures and animations for multi-channel images with a focus on microscopy.
Project description
microfilm
This package is a collection of tools to display and analyze 2D and 2D time-lapse microscopy images. In particular it makes it straightforward to create figures containing multi-channel images represented in a composite color mode as done in the popular image processing software Fiji. It also allows to easily complete such figures with standard annotations like labels and scale bars. In case of time-lapse data, the figures are turned into animations which can be interactively browsed from a Jupyter notebook, saved in standard movie formats (mp4, gif etc.) and completed with time counters. Finally, figures and animations can easily be combined into larger panels. These main functionalities are provided by the microfilm.microplot and microfilm.microanim modules.
Following the model of seaborn, microfilm is entirely based on Matplotlib and tries to provide good defaults to produce good microcopy figures out-of-the-box. It however also offers complete access to the Matplotlib structures like axis and figures underlying the microfilm objects, allowing thus for the creation of arbitrarily complex plots.
Installation
You can install this package directly from Github using:
pip install microfilm
To test the package via the Jupyter interface and the notebooks available here you can create a conda environment using the environment.yml file:
conda env create -f environment.yml
Simple plot
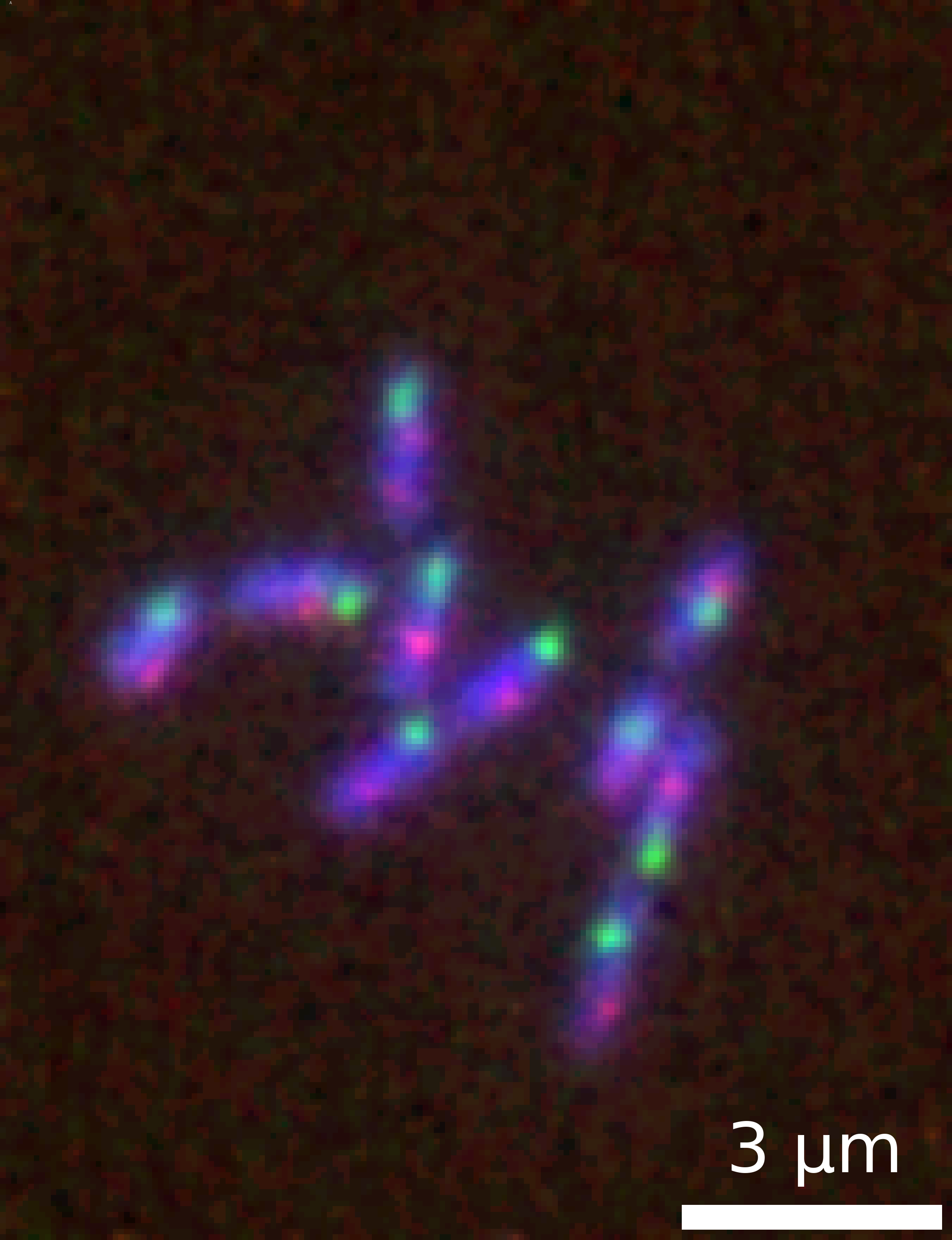
It is straightforward to create a ready-to-use plot of a multi-channel image dataset. In the following code snippet, we load a Numpy array of a multi-channel time-lapse dataset with shape CTXY (three channels). The figure below showing the time-point t=10 is generated in a single command with a few options and saved as a png:
import numpy as np
import skimage.io
from microfilm.microplot import microshow
image = skimage.io.imread('../demodata/coli_nucl_ori_ter.tif')
time = 10
microim = microshow(images=image[:, time, :, :], fig_scaling=5,
cmaps=['pure_blue','pure_red', 'pure_green'],
unit='um', scalebar_size_in_units=3, scalebar_unit_per_pix=0.065, scalebar_text_centered=True, scalebar_font_size=0.04,label_text='A', label_font_size=0.04)
microim.savefig('../illustrations/composite.png', bbox_inches = 'tight', pad_inches = 0, dpi=600)
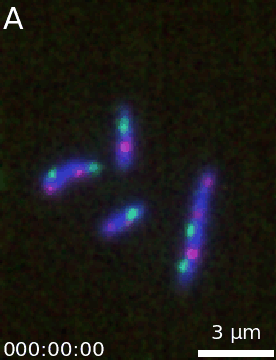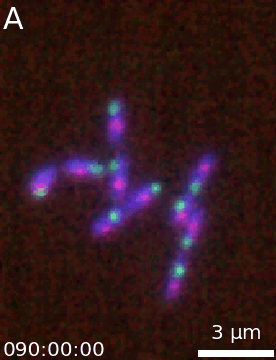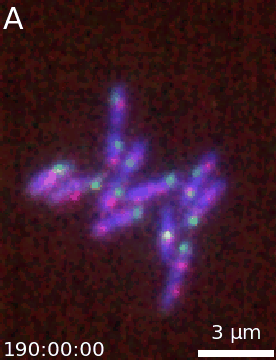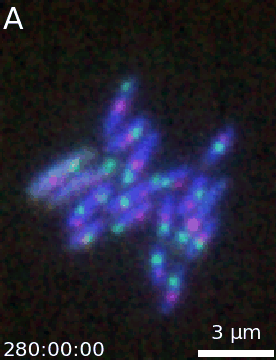
Animation
It is then easy to extend a simple figure into an animation as both objects take the same options. Additionally, a time-stamp can be added to the animation. This code generates the movie visible below:
import numpy as np
import skimage.io
from microfilm.microanim import Microanim
image = skimage.io.imread('../demodata/coli_nucl_ori_ter.tif')
microanim = Microanim(data=image, cmaps=['pure_blue','pure_red', 'pure_green'], fig_scaling=5,
unit='um', scalebar_size_in_units=3, scalebar_unit_per_pix=0.065,
scalebar_font_size=0.04)
microanim.add_label('A', label_font_size=30)
microanim.add_time_stamp('T', 10, location='lower left', timestamp_size=20)
microanim.save_movie('../illustrations/composite_movie.gif', fps=15)
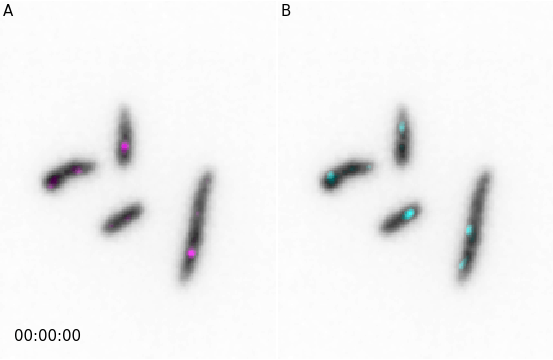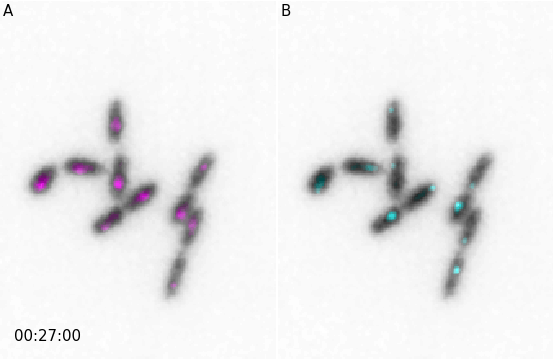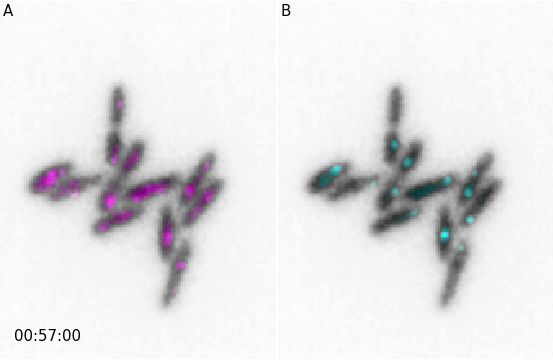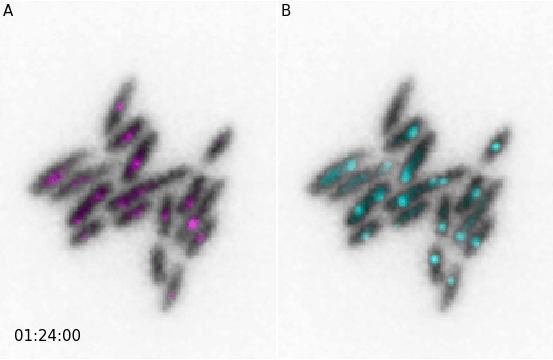
Panels
Both simple figures and animations can be combined into larger panels via the microplot.Micropanel and microanim.Microanimpanel objects. For example we can first create two figures microim1 and microim2 and then combine them into micropanel:
from microfilm import microplot
import skimage.io
image = skimage.io.imread('../demodata/coli_nucl_ori_ter.tif')
microim1 = microplot.microshow(images=[image[0, 10, :, :], image[1, 10, :, :]],
cmaps=['gray', 'pure_magenta'], flip_map=[True, False],
label_text='A', label_color='black')
microim2 = microplot.microshow(images=[image[0, 10, :, :], image[2, 10, :, :]],
cmaps=['gray', 'pure_cyan'], flip_map=[True, False],
label_text='B', label_color='black')
micropanel = microplot.Micropanel(rows=1, cols=2, figsize=[4,3])
micropanel.add_element(pos=[0,0], microim=microim1)
micropanel.add_element(pos=[0,1], microim=microim2)
micropanel.savefig('../illustrations/panel.png', bbox_inches = 'tight', pad_inches = 0, dpi=600)
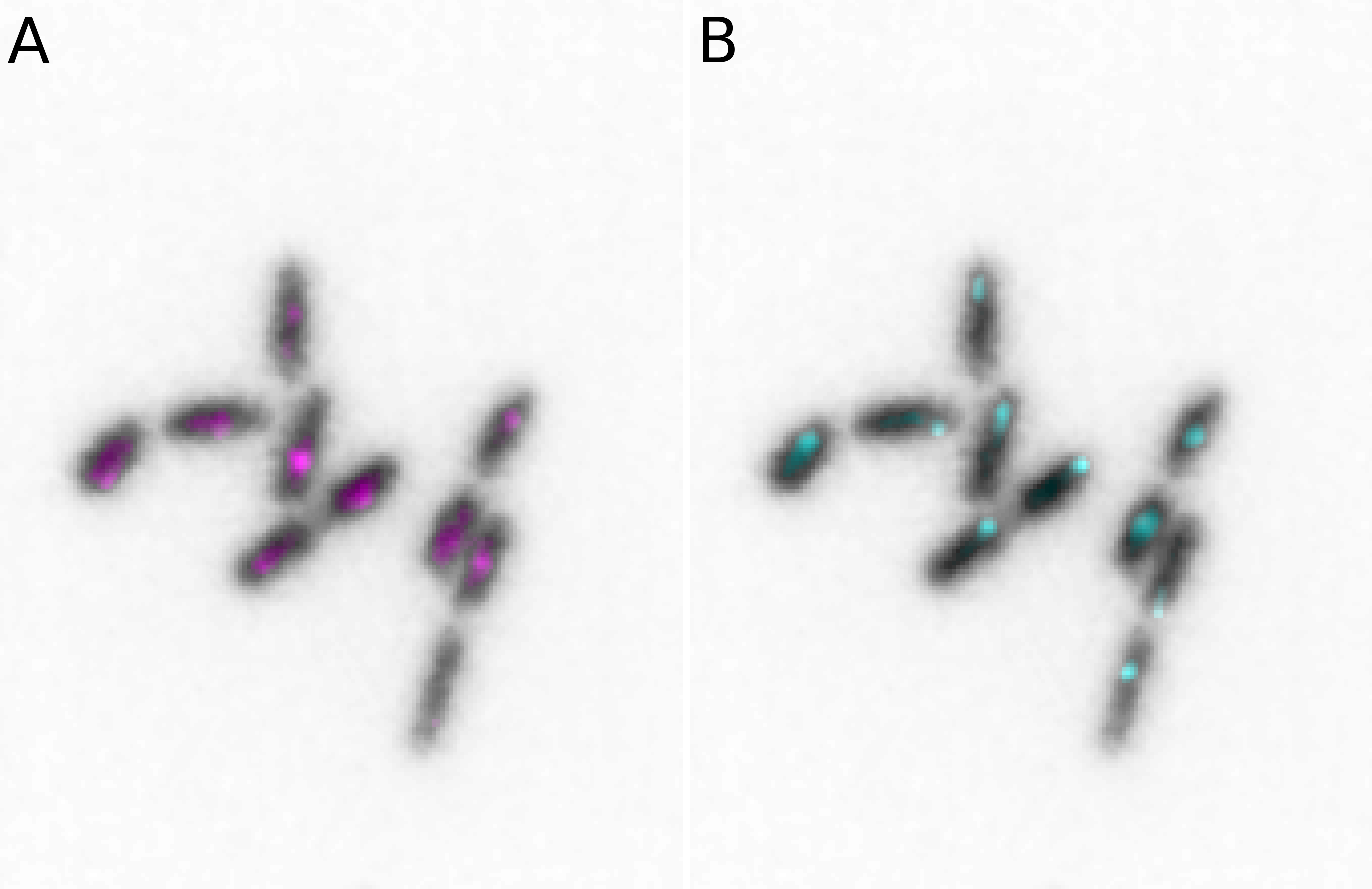
And similarly for animations:
from microfilm import microanim
import skimage.io
image = skimage.io.imread('../demodata/coli_nucl_ori_ter.tif')
microanim1 = microanim.Microanim(data=image[[0,1],::], cmaps=['gray', 'pure_magenta'],
flip_map=[True, False], label_text='A', label_color='black')
microanim2 = microanim.Microanim(data=image[[0,2],::], cmaps=['gray', 'pure_cyan'],
flip_map=[True, False], label_text='B', label_color='black')
microanim1.add_time_stamp(unit='T', unit_per_frame='3', location='lower-right', timestamp_color='black')
animpanel = microanim.Microanimpanel(rows=1, cols=2, figsize=[4,3])
animpanel.add_element(pos=[0,0], microanim=microanim1)
animpanel.add_element(pos=[0,1], microanim=microanim2)
animpanel.save_movie('../illustrations/panel.gif')
Additional functionalities
In addition to these main plotting capabilities, the packages also offers:
microfilm.colorify: a series of utility functions used by the main functions to create the composite color images. It contains functions to create colormaps, to turn 2D arrays into 3D-RGB arrays with appropriate colormaps etc.microfilm.dataset: a module offering a simple common data structure to handle multi-channel time-lapse data from multipage tiffs, series of tiff files, Nikon ND2 files, H5 and Numpy arrays. Requirement to use this module are at the moment very constrained (e.g. dimension order of Numpy arrays, name of H5 content etc.) but might evolve in the future.
Authors
This package has been created by Guillaume Witz, Microscopy Imaging Center and Science IT Support, University of Bern.
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
File details
Details for the file microfilm-0.0.8.tar.gz.
File metadata
- Download URL: microfilm-0.0.8.tar.gz
- Upload date:
- Size: 72.6 MB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/3.4.1 importlib_metadata/4.6.1 pkginfo/1.7.0 requests/2.25.1 requests-toolbelt/0.9.1 tqdm/4.61.2 CPython/3.9.6
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 3b27b909f48806e3ae7ba7cb19a0d5cce85e3103fcf9396f068094d1aea295a8 |
|
| MD5 | 6a52c64bf3803ed53fd292f06701ee1e |
|
| BLAKE2b-256 | 32f9f68ba8a7ea2603196b3bd1d8834ee26744033867e14993e15f3f122399ca |
File details
Details for the file microfilm-0.0.8-py3-none-any.whl.
File metadata
- Download URL: microfilm-0.0.8-py3-none-any.whl
- Upload date:
- Size: 21.6 kB
- Tags: Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/3.4.1 importlib_metadata/4.6.1 pkginfo/1.7.0 requests/2.25.1 requests-toolbelt/0.9.1 tqdm/4.61.2 CPython/3.9.6
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 007ed6ad15c0d4475f830f356a4baff226954e155e1dc6725018e81904e5acba |
|
| MD5 | 0dbe1c08cf641332db14848bf4c32e1a |
|
| BLAKE2b-256 | 3b95bb2a257b6abcff7746403b2a87a94279c12335d866991d3205c0dd0fd277 |

















