A pipeline for processing open medical examier's data using GitHub Actions CI/CD.
Project description
Medical Examiner Open Data Pipeline

This repository contains the code for the Medical Examiner Open Data Pipeline.
We currently fetch data from the following sources:
- Cook County Medical Examiner's Archives
- San Diego Medical Examiner's Office
- Milwaukee County Medical Examiner's Office
- Connecticut (State) Accidental Drug Deaths
- Santa Clara County Medical Examiner's Office
- Sacramento County Medical Examiner's Office
- Pima County Medical Examiner's Office
The results of this data are used in various other analysis here on GitHub:
- Cook County
- Where we add geospatial data to the Cook County data
- This was excluded from this automated pipeline due to specific requirements for the data for only Cook County
- Where we add geospatial data to the Cook County data
Getting Started
This repo exists mainly to take advantage of GitHub actions for automation.
The actions workflow is located in .github/workflows/pipeline.yml and is triggered weekly or manually.
This workflow fetches data from the configured data sources inside config.json,
geocodes addresses (when available) using ArcGIS, extracts drugs using the drug extraction toolbox
and then compiles and zips up the results into the GitHub Releases page.
The data is then available for download from the releases page page.
Further, the entire workflow effectively runs a series of commands using the CLI application opendata-pipeline which is located in the src directory.
This is also available via a docker image hosted on ghcr.io. The benefits of using the CLI via a docker image is that you don't have to have Python or the drug toolbox on your local machine 🙂.
We utilize async methods to speed up the large number of web requests we make to the data sources.
It is important to regularly fetch/pull from this repo to maintain an updated
config.json
We currently do not guarantee Windows support unfortunately. If you want to help make that a reality, please submit a new Pull Request
There is further API-documentation available on the GitHub Pages website for this repo if you want to interact with the CLI.
I would recommend using the docker image as it is easier to use and always referring to the CLI --help for more information.
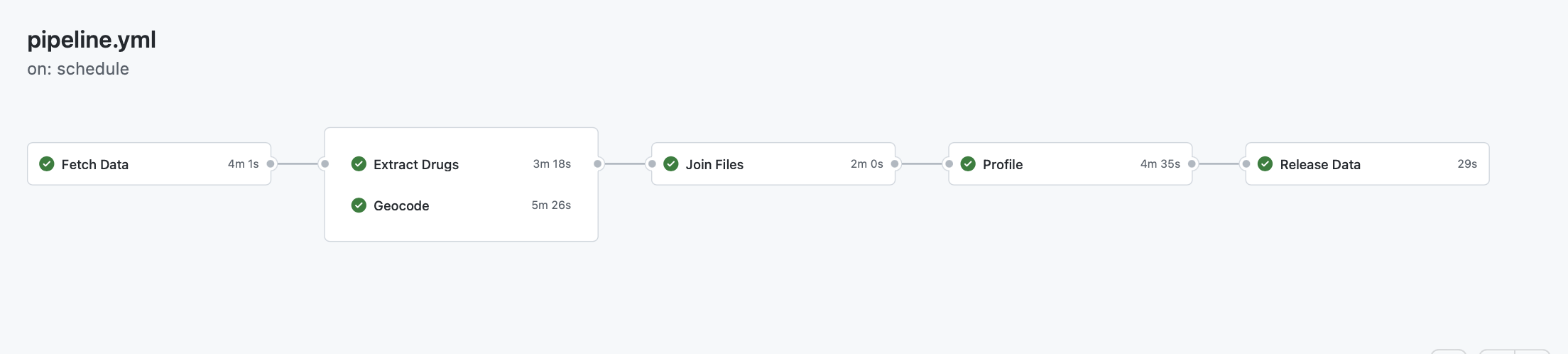
Workflow
The workflow can best be described by looking at the pipeline.yml file.
Data Enhancements
The following table shows the fields that we add to the original datafiles:
| Column Name | Description |
|---|---|
CaseIdentifier |
A unique identifier across all the datasets. |
death_day |
Day of the Month death occurred |
death_month |
Month Name death occurred |
death_month_num |
Month Number death occurred |
death_year |
Year death occurred |
death_day_of_week |
Day of week death occurred. Starting with 0 on Monday. Weekends are 5 (Saturday) & 6 (Sunday). |
death_day_is_weekend |
Death occurred on weekend day |
death_day_week_of_year |
Week of the year (of 52) that death occurred |
geocoded_latitude |
Geocoded latitude. |
geocoded_longitude |
Geocoded longitude. |
geocoded_score |
Confidence of geocoding. 70-100. |
geocoded_address |
The address that the geocoded results correspond to. Not the address provided to the geocoder. |
Drug Columns
In addition to providing the extracted drugs as a separate file in each release, we also convert this data to wide-form for each dataset. This adds the following columns in the subsequent pattern:
| Column Name/Pattern | Description |
|---|---|
*_1 |
* drug found in first search column provided in drug configuration |
*_2 |
* drug found in second search column provided in drug configuration |
*_meta |
Drug of * category/class found in this record across any search column. |
Requirements
uv
Installation
To install the python cli I recommend using uv.
uvx opendata-pipeline
To install the docker image, you can use the following command:
docker pull ghcr.io/uk-ipop/opendata-pipeline:latest
Usage
Usage is very similar to any other command line application. The most important thing is to follow the workflow defined above.
Contributing
Pull requests are welcome. For major changes, please open an issue first to discuss what you would like to change.
Help me write some tests!
License
BibTex Citation
If you use this software or the enhanced data, please cite this repository:
@software{Anthony_Medical_Examiner_OpenData_2022,
author = {Anthony, Nicholas},
month = {9},
title = {{Medical Examiner OpenData Pipeline}},
url = {https://github.com/UK-IPOP/open-data-pipeline},
version = {0.2.1},
year = {2022}
}
Thank you.
Project details
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
File details
Details for the file opendata_pipeline-0.3.0.tar.gz.
File metadata
- Download URL: opendata_pipeline-0.3.0.tar.gz
- Upload date:
- Size: 122.9 kB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: uv/0.5.1
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 10a93ccdcdc9ab59847abd16d7cf84ad4b9bb0b8498abf7af1552053c667a47c |
|
| MD5 | 43a6e7663756c8d2c951b455f379729b |
|
| BLAKE2b-256 | 6597439ff68f7a225a7a219e4b37e4c7c46e0594793cf31ba60ba1fc87784439 |
File details
Details for the file opendata_pipeline-0.3.0-py3-none-any.whl.
File metadata
- Download URL: opendata_pipeline-0.3.0-py3-none-any.whl
- Upload date:
- Size: 34.2 kB
- Tags: Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: uv/0.5.1
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 59315156aa480edeb1ff65426e0f3362059d8e97619d7213aa7865454444c643 |
|
| MD5 | 50ca915db326cb79a4b20278bfca8d47 |
|
| BLAKE2b-256 | 76a42ec58b71f3992158e0258102aca87f51029a01cdadc776c071a57c6428cb |













