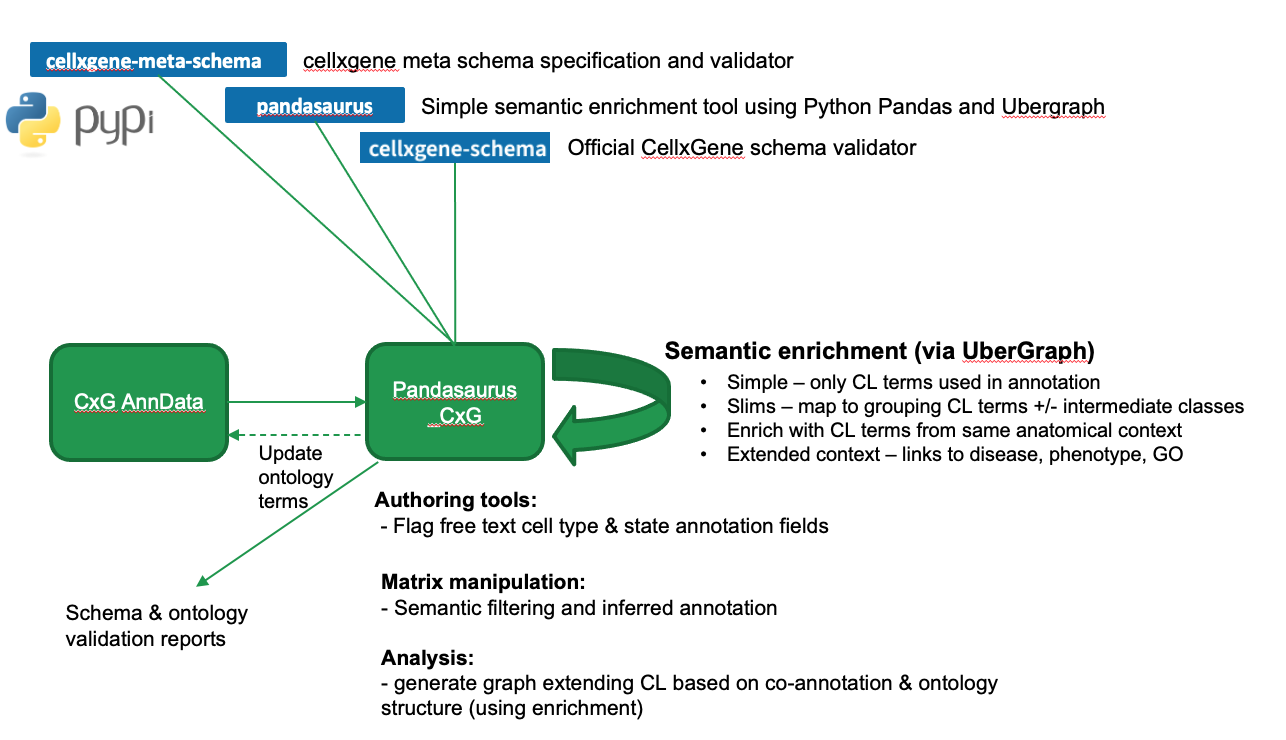
Ontology enrichment tool for CxG standard AnnData files.
Project description
pandasaurus_cxg
STATUS: early Beta
A library for retrieving and leveraging the semantic context of ontology annotation in CxG standard AnnData files.
Slide summarising intended functionality
Installation
Available on PyPi
$ pip3 install pandasaurus_cxg
Detailed installation guide for pygraphviz issue
During package installation, sometimes the pygraphviz package installation is failing on macOS due to Graphviz may be installed in a location that is not on the default search path. In this case, it may be necessary to manually specify the path to the graphviz include and/or library directories. To do that use following instructions and install pygraphviz manually.
brew install graphviz
export CFLAGS="-I$(brew --prefix graphviz)/include"
export LDFLAGS="-L$(brew --prefix graphviz)/lib"
pip install pygraphviz
Usage
The AnndataEnricher and AnndataAnalyzer classes can be used both individually and in conjunction with the AnndataEnrichmentAnalyzer wrapper class. The AnndataEnrichmentAnalyzer class serves as a convenient way to leverage the functionalities of both AnndataEnricher and AnndataAnalyzer.
Using AnndataEnricher and AnndataAnalyzer Individually
You can use the AnndataEnricher and AnndataAnalyzer classes separately to perform specific tasks on your data. For instance, AnndataEnricher facilitates data enrichment, while AnndataAnalyzer provides various analysis tools for Anndata objects.
from pandasaurus_cxg.anndata_enricher import AnndataEnricher
ade = AnndataEnricher.from_file_path("test/data/modified_human_kidney.h5ad")
ade.simple_enrichment()
ade.minimal_slim_enrichment(["blood_and_immune_upper_slim"])
from pandasaurus_cxg.anndata_analyzer import AnndataAnalyzer
ada = AnndataAnalyzer.from_file_path("./immune_example.h5ad", author_cell_type_list = ['subclass.full', 'subclass.l3', 'subclass.l2', 'subclass.l1', 'class', 'author_cell_type'])
ada.co_annotation_report()
Using AnndataEnrichmentAnalyzer Wrapper
The AnndataEnrichmentAnalyzer class wraps the functionality of both AnndataEnricher and AnndataAnalyzer, offering a seamless way to perform enrichment and analysis in one go.
from pandasaurus_cxg.enrichment_analysis import AnndataEnrichmentAnalyzer
from pandasaurus_cxg.graph_generator.graph_generator import GraphGenerator
aea = AnndataEnrichmentAnalyzer("test/data/modified_human_kidney.h5ad")
aea.contextual_slim_enrichment()
aea.co_annotation_report()
gg = GraphGenerator(aea)
gg.generate_rdf_graph()
gg.set_label_adding_priority(["class", "cell_type", "subclass.l1", "subclass.l1", "subclass.full", "subclass.l2", "subclass.l3"])
gg.add_label_to_terms()
gg.enrich_rdf_graph()
gg.save_rdf_graph(file_name="kidney_new", _format="ttl")
More examples and detailed explanation can be found in jupyter notebook given in Snippets
Snippets
https://github.com/INCATools/pandasaurus_cxg/blob/main/walkthrough.ipynb
Library Documentation
https://incatools.github.io/pandasaurus_cxg/
Roadmap
https://github.com/INCATools/pandasaurus_cxg/blob/main/ROADMAP.md
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
Hashes for pandasaurus_cxg-0.1.24-py3-none-any.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 098e11432d99a0af29c757cae06a93c4f4f7101ea3dfd8709f6e421af7473d83 |
|
| MD5 | 580bf91222ce6dd98a49d44baa58ca58 |
|
| BLAKE2b-256 | 1c3faf27828c1caf06cfbc199788578f9e126df7891aeda4d2ff01a8780b3485 |