REvolutionH-tl: Reconstruction of Evolutionary Histories tool
Project description
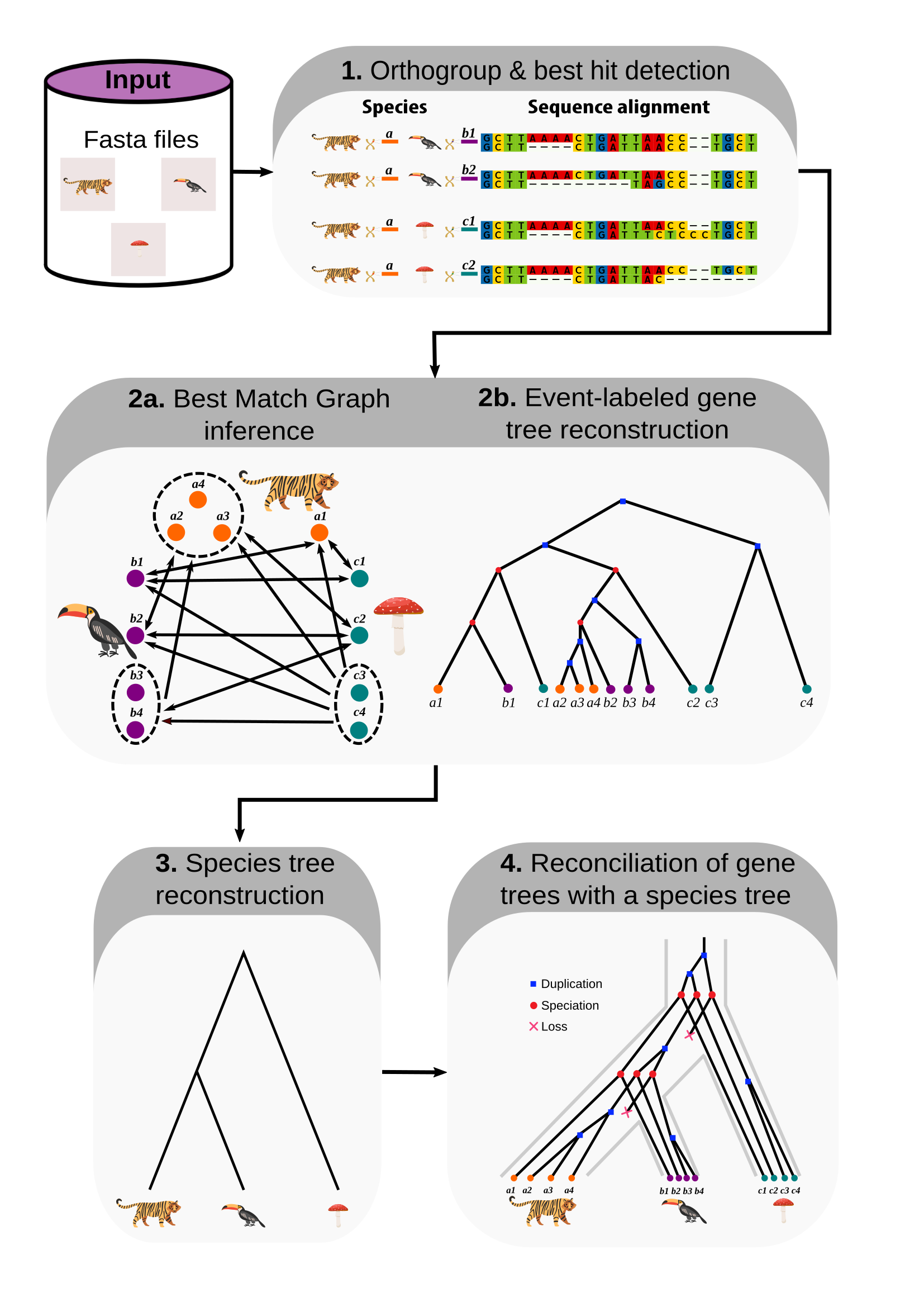
Bioinformatics tool for the reconstruction of evolutionary histories. Input: fasta files or sequence alignment hits, Output: event-labeled gene trees and reconciliations.
Bioinformatics & complex networks lab
- José Antonio Ramírez-Rafael [jose.ramirezra@cinvestav.mx]
- Maribel Hernandez-Rosales [maribel.hr@cinvestav.mx ]
Install
pip install revolutionh-tl
Requirements
For sequence alignments using revolutionhtl: Diamond
Usage
python -m revolutionhtl <arguments>
Pipeline
- Orthogroup & best hit selection. Input: alignment hits (generate this using
revolutionhtl.diamond) . - Orthology and gene tree reconstruction. Input: best hits (generate this at step 1).
- Species tree reconstruction. Input: gene trees (generate this at step 2).
- Tree reconciliation. Input: gene and species trees (generate this at steps 2 and 3).
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
revolutionhtl-1.0.1.tar.gz
(31.3 kB
view hashes)
Built Distribution
Close
Hashes for revolutionhtl-1.0.1-py3-none-any.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 24f0a29614b93bd25c5ccb32e094a2a275b6d852442d3edce6fd0655eb9e261c |
|
| MD5 | b5f839bbb8a67503289a9247be47049f |
|
| BLAKE2b-256 | 5da67f9305828e5336a1911973b834c44e019f6871570c3864ca18440a8f8cf1 |












