REvolutionH-tl: Reconstruction of Evolutionary Histories tool
Project description
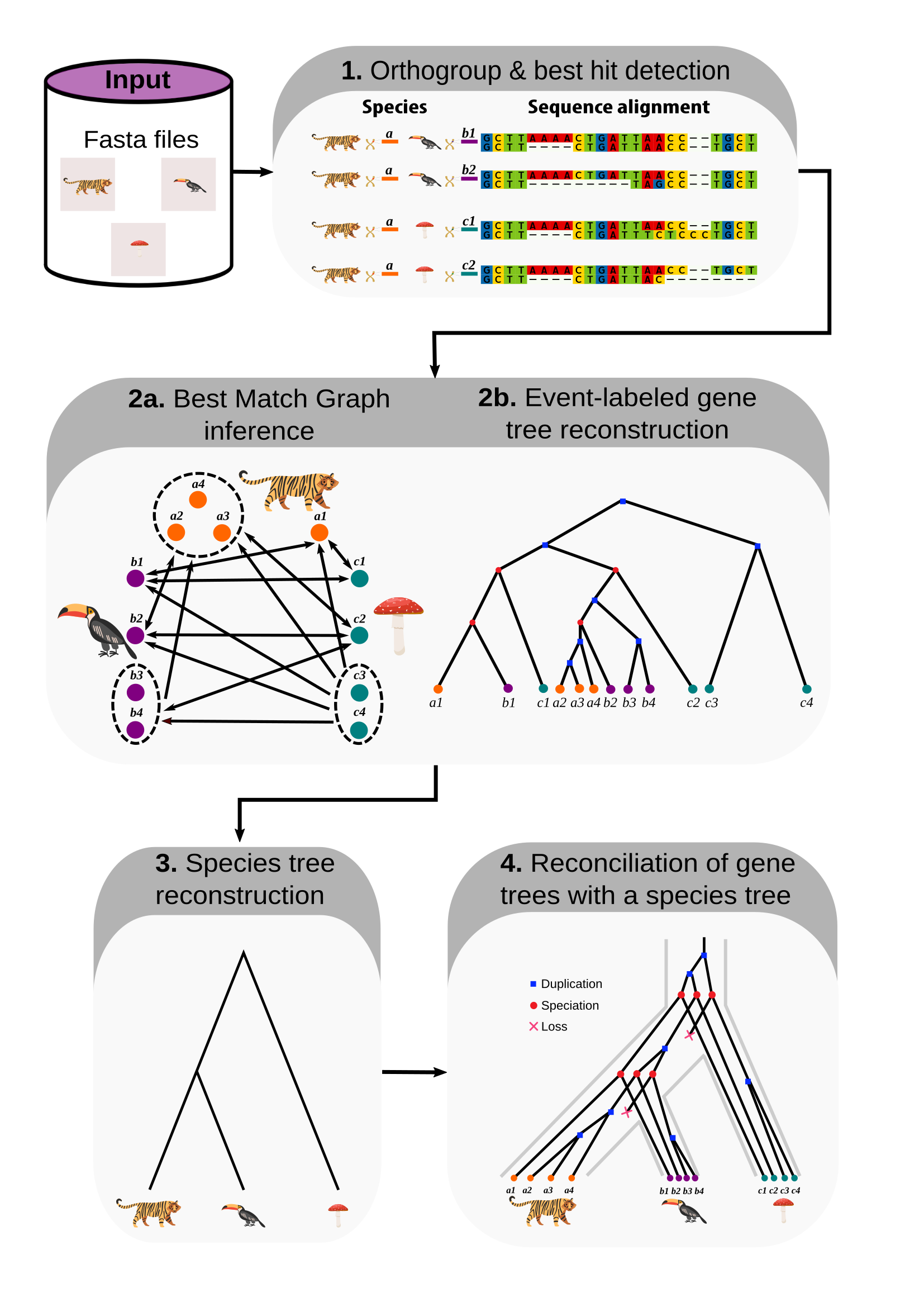
Bioinformatics tool for the reconstruction of evolutionary histories. Input: fasta files or sequence alignment hits, Output: orthology. event-labeled gene trees, and reconciliations.
Bioinformatics & complex networks lab
- José Antonio Ramírez-Rafael [jose.ramirezra@cinvestav.mx]
- Maribel Hernandez-Rosales [maribel.hr@cinvestav.mx ]
Steps
- Orthogroup & best hit selection. Input: alignment hits (generate this using
revolutionhtl.diamond) . - Orthology and gene tree reconstruction. Input: best hits (generate this at step 1).
- Species tree reconstruction. Input: gene trees (generate this at step 2).
- Tree reconciliation. Input: gene and species trees (generate this at steps 2 and 3).
Install
pip install revolutionhtl
Requirements
If you want to run sequence alignments using revolutionhtl, then install Diamond.
Usage
python -m revolutionhtl <arguments>
The arguments specify steps to run and parameters.
Specify steps and input data (Click to expand)
- -h --help show this help message and exit-steps [int ...] List of steps to run (default: 1 2 3 4).
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
revolutionhtl-1.0.9.tar.gz
(35.3 kB
view hashes)
Built Distribution
Close
Hashes for revolutionhtl-1.0.9-py3-none-any.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | f4a2f57928ce831b2cd69e94a977d3364ab30d7093d8cfb63e077f5703df4e69 |
|
| MD5 | 0fb350ebee3dec15de1a6f82fd166854 |
|
| BLAKE2b-256 | f76148f33885e92482f11dea6540e4eef3170c137f075da73b8d0eedb371f469 |












