TemporAI: ML-centric Toolkit for Medical Time Series
Project description
 TemporAI
TemporAI
⚗️ Status: This project is still in alpha, and the API may change without warning.
📃 Overview
TemporAI is a Machine Learning-centric time-series library for medicine. The tasks that are currently of focus in TemporAI are: time-to-event (survival) analysis with time-series data, treatment effects (causal inference) over time, and time-series prediction. Data preprocessing methods, including missing value imputation for static and temporal covariates, are provided. AutoML tools for hyperparameter tuning and pipeline selection are also available.
How is TemporAI unique?
- 🏥 Medicine-first: Focused on use cases for medicine and healthcare, such as temporal treatment effects, survival analysis over time, imputation methods, models with built-in and post-hoc interpretability, ... See methods.
- 🏗️ Fast prototyping: A plugin design allowing for on-the-fly integration of new methods by the users.
- 🚀 From research to practice: Relevant novel models from research community adapted for practical use.
- 🌍 A healthcare ecosystem vision: A range of interactive demonstration apps, new medical problem settings, interpretability tools, data-centric tools etc. are planned.
Key concepts
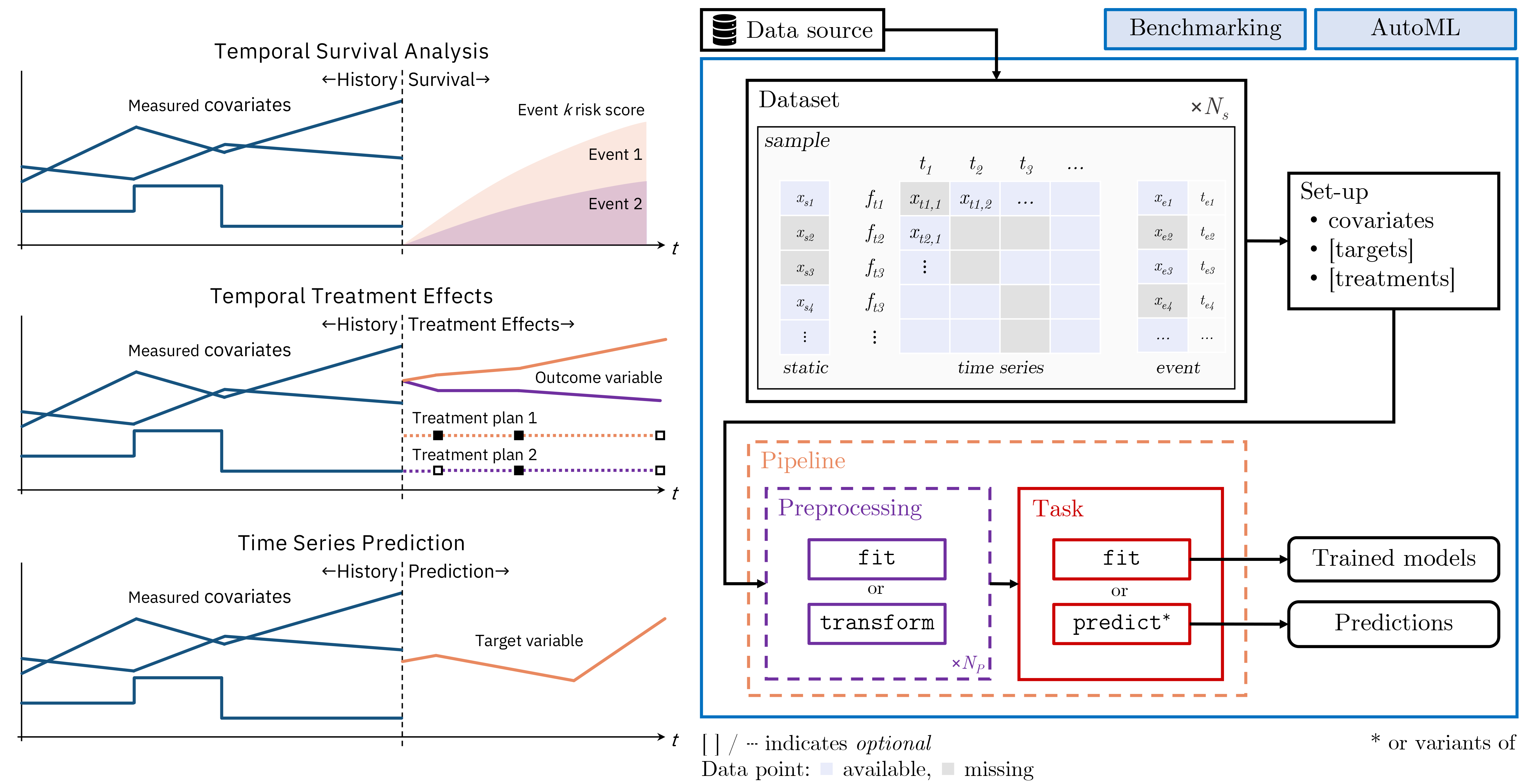
🚀 Installation
Instal with pip
From the Python Package Index (PyPI):
$ pip install temporai
Or from source:
$ git clone https://github.com/vanderschaarlab/temporai.git
$ cd temporai
$ pip install .
Install in a conda environment
While have not yet published TemporAI on conda-forge, you can still install TemporAI in your conda environment using pip as follows:
Create and activate conda environment as normal:
$ conda create -n <my_environment>
$ conda activate <my_environment>
Then install inside your conda environment with pip:
$ pip install temporai
💥 Sample Usage
- List the available plugins
from tempor import plugin_loader
print(plugin_loader.list())
- Use a time-to-event (survival) analysis model
from tempor import plugin_loader
# Load a time-to-event dataset:
dataset = plugin_loader.get("time_to_event.pbc", plugin_type="datasource").load()
# Initialize the model:
model = plugin_loader.get("time_to_event.dynamic_deephit")
# Train:
model.fit(dataset)
# Make risk predictions:
prediction = model.predict(dataset, horizons=[0.25, 0.50, 0.75])
- Use a temporal treatment effects model
import numpy as np
from tempor import plugin_loader
# Load a dataset with temporal treatments and outcomes:
dataset = plugin_loader.get(
"treatments.temporal.dummy_treatments",
plugin_type="datasource",
temporal_covariates_missing_prob=0.0,
temporal_treatments_n_features=1,
temporal_treatments_n_categories=2,
).load()
# Initialize the model:
model = plugin_loader.get("treatments.temporal.regression.crn_regressor", epochs=20)
# Train:
model.fit(dataset)
# Define target variable horizons for each sample:
horizons = [
tc.time_indexes()[0][len(tc.time_indexes()[0]) // 2 :] for tc in dataset.time_series
]
# Define treatment scenarios for each sample:
treatment_scenarios = [
[np.asarray([1] * len(h)), np.asarray([0] * len(h))] for h in horizons
]
# Predict counterfactuals:
counterfactuals = model.predict_counterfactuals(
dataset,
horizons=horizons,
treatment_scenarios=treatment_scenarios,
)
- Use a missing data imputer
from tempor import plugin_loader
dataset = plugin_loader.get(
"prediction.one_off.sine", plugin_type="datasource", with_missing=True
).load()
static_data_n_missing = dataset.static.dataframe().isna().sum().sum()
temporal_data_n_missing = dataset.time_series.dataframe().isna().sum().sum()
print(static_data_n_missing, temporal_data_n_missing)
assert static_data_n_missing > 0
assert temporal_data_n_missing > 0
# Initialize the model:
model = plugin_loader.get("preprocessing.imputation.temporal.bfill")
# Train:
model.fit(dataset)
# Impute:
imputed = model.transform(dataset)
temporal_data_n_missing = imputed.time_series.dataframe().isna().sum().sum()
print(static_data_n_missing, temporal_data_n_missing)
assert temporal_data_n_missing == 0
- Use a one-off classifier (prediction)
from tempor import plugin_loader
dataset = plugin_loader.get("prediction.one_off.sine", plugin_type="datasource").load()
# Initialize the model:
model = plugin_loader.get("prediction.one_off.classification.nn_classifier", n_iter=50)
# Train:
model.fit(dataset)
# Predict:
prediction = model.predict(dataset)
- Use a temporal regressor (forecasting)
from tempor import plugin_loader
# Load a dataset with temporal targets.
dataset = plugin_loader.get(
"prediction.temporal.dummy_prediction",
plugin_type="datasource",
temporal_covariates_missing_prob=0.0,
).load()
# Initialize the model:
model = plugin_loader.get("prediction.temporal.regression.seq2seq_regressor", epochs=10)
# Train:
model.fit(dataset)
# Predict:
prediction = model.predict(dataset, n_future_steps=5)
- Benchmark models, time-to-event task
from tempor.benchmarks import benchmark_models
from tempor import plugin_loader
from tempor.methods.pipeline import pipeline
testcases = [
(
"pipeline1",
pipeline(
[
"preprocessing.scaling.temporal.ts_minmax_scaler",
"time_to_event.dynamic_deephit",
]
)({"ts_coxph": {"n_iter": 100}}),
),
(
"plugin1",
plugin_loader.get("time_to_event.dynamic_deephit", n_iter=100),
),
(
"plugin2",
plugin_loader.get("time_to_event.ts_coxph", n_iter=100),
),
]
dataset = plugin_loader.get("time_to_event.pbc", plugin_type="datasource").load()
aggr_score, per_test_score = benchmark_models(
task_type="time_to_event",
tests=testcases,
data=dataset,
n_splits=2,
random_state=0,
horizons=[2.0, 4.0, 6.0],
)
print(aggr_score)
- Serialization
from tempor.utils.serialization import load, save
from tempor import plugin_loader
# Initialize the model:
model = plugin_loader.get("prediction.one_off.classification.nn_classifier", n_iter=50)
buff = save(model) # Save model to bytes.
reloaded = load(buff) # Reload model.
# `save_to_file`, `load_from_file` also available in the serialization module.
- AutoML - search for the best pipeline for your task
from tempor.automl.seeker import PipelineSeeker
dataset = plugin_loader.get("prediction.one_off.sine", plugin_type="datasource").load()
# Specify the AutoML pipeline seeker for the task of your choice, providing candidate methods,
# metric, preprocessing steps etc.
seeker = PipelineSeeker(
study_name="my_automl_study",
task_type="prediction.one_off.classification",
estimator_names=[
"cde_classifier",
"ode_classifier",
"nn_classifier",
],
metric="aucroc",
dataset=dataset,
return_top_k=3,
num_iter=100,
tuner_type="bayesian",
static_imputers=["static_tabular_imputer"],
static_scalers=[],
temporal_imputers=["ffill", "bfill"],
temporal_scalers=["ts_minmax_scaler"],
)
# The search will return the best pipelines.
best_pipelines, best_scores = seeker.search() # doctest: +SKIP
📖 Tutorials
Data
User Guide
- Plugins
- Imputation
- Scaling
- Prediction
- Time-to-event Analysis
- Treatment Effects
- Pipeline
- Benchmarks
- AutoML
Extending TemporAI
- Writing a Custom Method Plugin
- Testing a Custom Method Plugin
- Writing a Custom Data Source Plugin
- Writing a Custom Metric Plugin
- Writing a Custom Data Format
📘 Documentation
See the full project documentation here.
🌍 TemporAI Ecosystem (Experimental)
We provide additional tools in the TemporAI ecosystem, which are in active development, and are currently (very) experimental. Suggestions and contributions are welcome!
These include:
temporai-clinic: A web app tool for interacting and visualising TemporAI models, data, and predictions.temporai-mivdp: A MIMIC-IV-Data-Pipeline adaptation for TemporAI.
🔑 Methods
Time-to-Event (survival) analysis over time
Risk estimation given event data (category: time_to_event)
| Name | Description | Reference |
|---|---|---|
dynamic_deephit |
Dynamic-DeepHit incorporates the available longitudinal data comprising various repeated measurements (rather than only the last available measurements) in order to issue dynamically updated survival predictions | Paper |
ts_coxph |
Create embeddings from the time series and use a CoxPH model for predicting the survival function | --- |
ts_xgb |
Create embeddings from the time series and use a SurvivalXGBoost model for predicting the survival function | --- |
Treatment effects
One-off
Treatment effects estimation where treatments are a one-off event.
- Regression on the outcomes (category:
treatments.one_off.regression)
| Name | Description | Reference |
|---|---|---|
synctwin_regressor |
SyncTwin is a treatment effect estimation method tailored for observational studies with longitudinal data, applied to the LIP setting: Longitudinal, Irregular and Point treatment. | Paper |
Temporal
Treatment effects estimation where treatments are temporal (time series).
- Classification on the outcomes (category:
treatments.temporal.classification)
| Name | Description | Reference |
|---|---|---|
crn_classifier |
The Counterfactual Recurrent Network (CRN), a sequence-to-sequence model that leverages the available patient observational data to estimate treatment effects over time. | Paper |
- Regression on the outcomes (category:
treatments.temporal.regression)
| Name | Description | Reference |
|---|---|---|
crn_regressor |
The Counterfactual Recurrent Network (CRN), a sequence-to-sequence model that leverages the available patient observational data to estimate treatment effects over time. | Paper |
Prediction
One-off
Prediction where targets are static.
- Classification (category:
prediction.one_off.classification)
| Name | Description | Reference |
|---|---|---|
nn_classifier |
Neural-net based classifier. Supports multiple recurrent models, like RNN, LSTM, Transformer etc. | --- |
ode_classifier |
Classifier based on ordinary differential equation (ODE) solvers. | --- |
cde_classifier |
Classifier based Neural Controlled Differential Equations for Irregular Time Series. | Paper |
laplace_ode_classifier |
Classifier based Inverse Laplace Transform (ILT) algorithms implemented in PyTorch. | Paper |
- Regression (category:
prediction.one_off.regression)
| Name | Description | Reference |
|---|---|---|
nn_regressor |
Neural-net based regressor. Supports multiple recurrent models, like RNN, LSTM, Transformer etc. | --- |
ode_regressor |
Regressor based on ordinary differential equation (ODE) solvers. | --- |
cde_regressor |
Regressor based Neural Controlled Differential Equations for Irregular Time Series. | Paper |
laplace_ode_regressor |
Regressor based Inverse Laplace Transform (ILT) algorithms implemented in PyTorch. | Paper |
Temporal
Prediction where targets are temporal (time series).
- Classification (category:
prediction.temporal.classification)
| Name | Description | Reference |
|---|---|---|
seq2seq_classifier |
Seq2Seq prediction, classification | --- |
- Regression (category:
prediction.temporal.regression)
| Name | Description | Reference |
|---|---|---|
seq2seq_regressor |
Seq2Seq prediction, regression | --- |
Preprocessing
Feature Encoding
- Static data (category:
preprocessing.encoding.static)
| Name | Description | Reference |
|---|---|---|
static_onehot_encoder |
One-hot encode categorical static features | --- |
- Temporal data (category:
preprocessing.encoding.temporal)
| Name | Description | Reference |
|---|---|---|
ts_onehot_encoder |
One-hot encode categorical time series features | --- |
Imputation
- Static data (category:
preprocessing.imputation.static)
| Name | Description | Reference |
|---|---|---|
static_tabular_imputer |
Use any method from HyperImpute (HyperImpute, Mean, Median, Most-frequent, MissForest, ICE, MICE, SoftImpute, EM, Sinkhorn, GAIN, MIRACLE, MIWAE) to impute the static data | Paper |
- Temporal data (category:
preprocessing.imputation.temporal)
| Name | Description | Reference |
|---|---|---|
ffill |
Propagate last valid observation forward to next valid | --- |
bfill |
Use next valid observation to fill gap | --- |
ts_tabular_imputer |
Use any method from HyperImpute (HyperImpute, Mean, Median, Most-frequent, MissForest, ICE, MICE, SoftImpute, EM, Sinkhorn, GAIN, MIRACLE, MIWAE) to impute the time series data | Paper |
Scaling
- Static data (category:
preprocessing.scaling.static)
| Name | Description | Reference |
|---|---|---|
static_standard_scaler |
Scale the static features using a StandardScaler | --- |
static_minmax_scaler |
Scale the static features using a MinMaxScaler | --- |
- Temporal data (category:
preprocessing.scaling.temporal)
| Name | Description | Reference |
|---|---|---|
ts_standard_scaler |
Scale the temporal features using a StandardScaler | --- |
ts_minmax_scaler |
Scale the temporal features using a MinMaxScaler | --- |
🔨 Tests and Development
Install the testing dependencies using:
pip install .[testing]
The tests can be executed using:
pytest -vsx
For local development, we recommend that you should install the [dev] extra, which includes [testing] and some additional dependencies:
pip install .[dev]
For development and contribution to TemporAI, see:
✍️ Citing
If you use this code, please cite the associated paper:
@article{saveliev2023temporai,
title={TemporAI: Facilitating Machine Learning Innovation in Time Domain Tasks for Medicine},
author={Saveliev, Evgeny S and van der Schaar, Mihaela},
journal={arXiv preprint arXiv:2301.12260},
year={2023}
}
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distributions
Built Distribution
File details
Details for the file temporai-0.0.3-py3-none-any.whl.
File metadata
- Download URL: temporai-0.0.3-py3-none-any.whl
- Upload date:
- Size: 236.6 kB
- Tags: Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/4.0.2 CPython/3.7.9
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | f05f88fa44a40724e57c1d29101869ba7453aae7374048f39decde8a96876516 |
|
| MD5 | 2d12b68a5552d3e85b32b0aaaba82430 |
|
| BLAKE2b-256 | 64def58349da04815bfb0466a0b6330af4e2497c06c9a0ebdb7aa00eda00e2b8 |























