Copy Number Variant Pathogenicity Classifier
Project description
CNVoyant
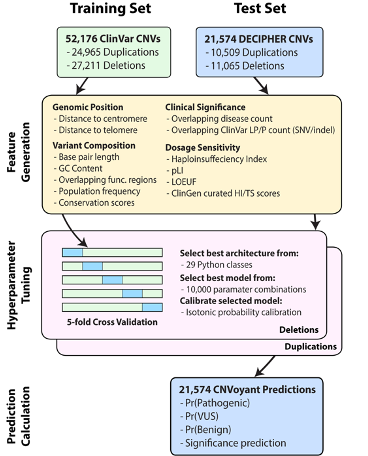
A collection of tools to annotate, predict clinical significance, and provide prediction explanations for Copy Number Variants (CNVs). Models were trained with the January 2023 version of ClinVar. Separate models were trained to predict deletion and duplication CNVs. To read more about features and benchmarking results, please see our recent publication in JOURNAL_LINK. Here is the graphical abstract of the project:
Dependencies
Python dependencies are handled via the anaconda package manager. The best way to create an environment with all needed dependencies is with conda or mamba (a conda wrapper that runs much faster). Create a new enviornment with CNVoyant with this command:
mamba create -n CNVoyant -c conda-forge -c bioconda python=3.10 schuetz.12::cnvoyant
Download Databases
CNVoyant requires ClinVar, conservation scores, functional region boundaries, gnomAD SV, and a GRCh38 reference genome to annotate inputted CNVs. To download these resources, a dependency directory must be specified and passed to the build_all method of the DependencyBuilder object.
from CNVoyant import DependencyBuilder
data_dir = '/path/to/cnvoyant_dependencies'
db = DependencyBuilder(data_dir)
db.build_all()
Build Features
CNVoyant features must be generated before predictions can be generated. Features can be generated by calling the get_features method from the FeatureBuilder object.
import pandas as pd
from CNVoyant import FeatureBuilder
# Create sample data
cnv_df = pd.DataFrame({
'CHROMOSOME': ['chr1','chr2','chr3','chr4','chr3'],
'START': [100000, 100000, 100000, 100000, 179197182],
'END': [200000, 200000, 200000, 200000, 179236784],
'CHANGE': ['DEL','DEL','DUP','DUP','DEL']
})
# Intialize CNVoyant FeatureBuilder instance
fb = FeatureBuilder(variant_df = cnv_df, data_dir = data_dir)
# Generate features
fb.get_features()
Generate Predictions
Pretrained models are available to generate predictions. Predictions can be generated by calling the predict method from the Classifier object.
from CNVoyant import Classifier
# Intialize CNVoyant Classifier instance
cl = Classifier(data_dir)
# Generate predictions
cnvoyant_preds = cl.predict(fb.feature_df)
Retrain CNVoyant Classifier
The CNVoyant models can be retrained to a specified set of variants, given that a label is available. Label values must be either 'Benign', 'VUS', or 'Pathogenic'. The name of the column header must be passed to the train method from the Classifier object.
from CNVoyant import FeatureBuilder, Classifier
# Sample data
cnv_train_df = pd.DataFrame({
'CHROMOSOME': ['chr1','chr2','chr3','chr4','chr3','chr8','chr8','chr8'],
'START': [100000,100000,100000,100000,179197182,60680919,38458191,37878455],
'END': [200000,200000,200000,200000,179236784,60738964,38470707,38884501],
'CHANGE': ['DUP','DEL','DUP','DUP','DEL','DEL','DUP','DUP'],
'LABEL': ['Benign','Benign','Benign','Benign','Pathogenic','VUS','VUS','Pathogenic']
})
# Intialize CNVoyant FeatureBuilder instance
fb_train = FeatureBuilder(variant_df = cnv_train_df, data_dir = data_dir)
# Generate features
fb_train.get_features()
# Intialize CNVoyant Classifier instance
cl_retrained = Classifier(data_dir)
# Retrain models
cl_retrained.train(fb_train.feature_df, label = 'LABEL')
# Generate predictions
cnvoyant_retrained_preds = cl_retrained.predict(fb.feature_df)
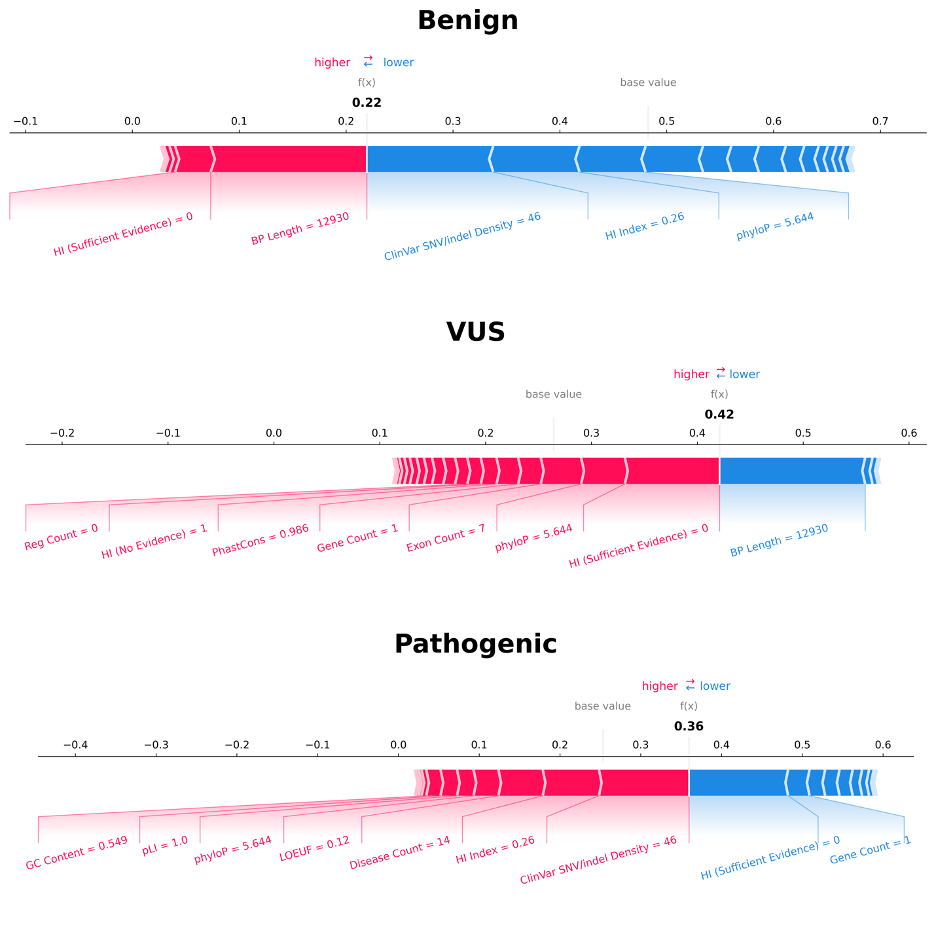
Generate CNVoyant Explanations
A key feature of CNVoyant is the ability to provide reasoning behind the provided clinical significance predictions. Explanations are provided via SHAP force plots, which indicate which features drove the prediction of each class for the provided CNV.
from CNVoyant import Explainer
cnv_coordinates = {
'CHROMOSOME': 'chr3',
'START': 179197182,
'END': 179236784,
'CHANGE': 'DEL'
}
expl = Explainer(
cnv_coordinates = cnv_coordinates,
output_dir = '/path/to/output',
classifier = cl
)
expl.explain()
The output looks like this:
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
Filter files by name, interpreter, ABI, and platform.
If you're not sure about the file name format, learn more about wheel file names.
Copy a direct link to the current filters
File details
Details for the file CNVoyant-1.1.6.tar.gz.
File metadata
- Download URL: CNVoyant-1.1.6.tar.gz
- Upload date:
- Size: 24.0 MB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/5.0.0 CPython/3.10.13
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
9da4b17305632f866da11188332401a897991d2c6615312240c615315fb26e7d
|
|
| MD5 |
c7574aaebbd9b7ce8ac3d58c2a865e10
|
|
| BLAKE2b-256 |
4d42667688b3a807503e1c3e5e504c94b04a872d770f1d067edcf80bd451c62c
|
File details
Details for the file CNVoyant-1.1.6-py3-none-any.whl.
File metadata
- Download URL: CNVoyant-1.1.6-py3-none-any.whl
- Upload date:
- Size: 24.1 MB
- Tags: Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/5.0.0 CPython/3.10.13
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
d7fb099ac4c5e4189aace3e86807d2ebec6335f9d26f858592a0b72d61184646
|
|
| MD5 |
1334369284831fb46e8352df0aefea41
|
|
| BLAKE2b-256 |
3dcc281d6599264668c25d83008572b2b99afc89ef80201e5d8e46a725cce25c
|