Cell Detection with PyTorch.
Project description
Cell Detection
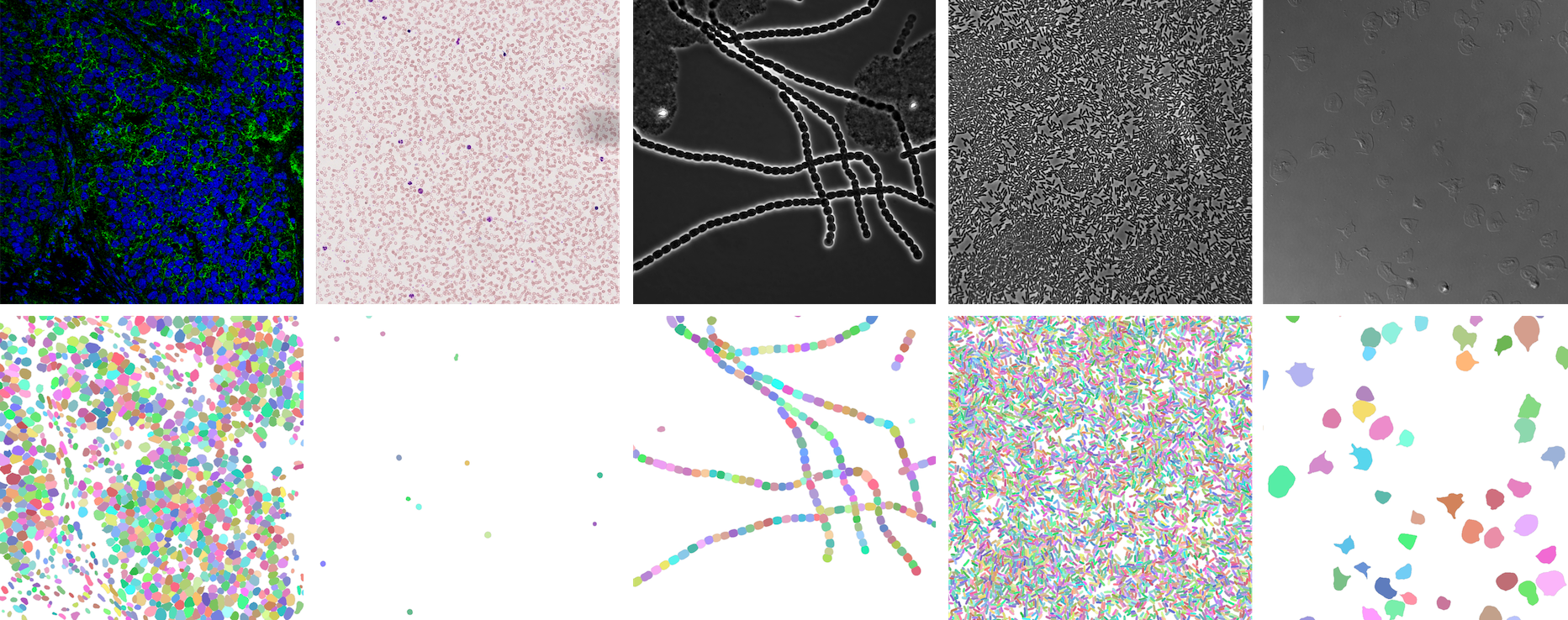
⭐ Showcase
NeurIPS 22 Cell Segmentation Competition
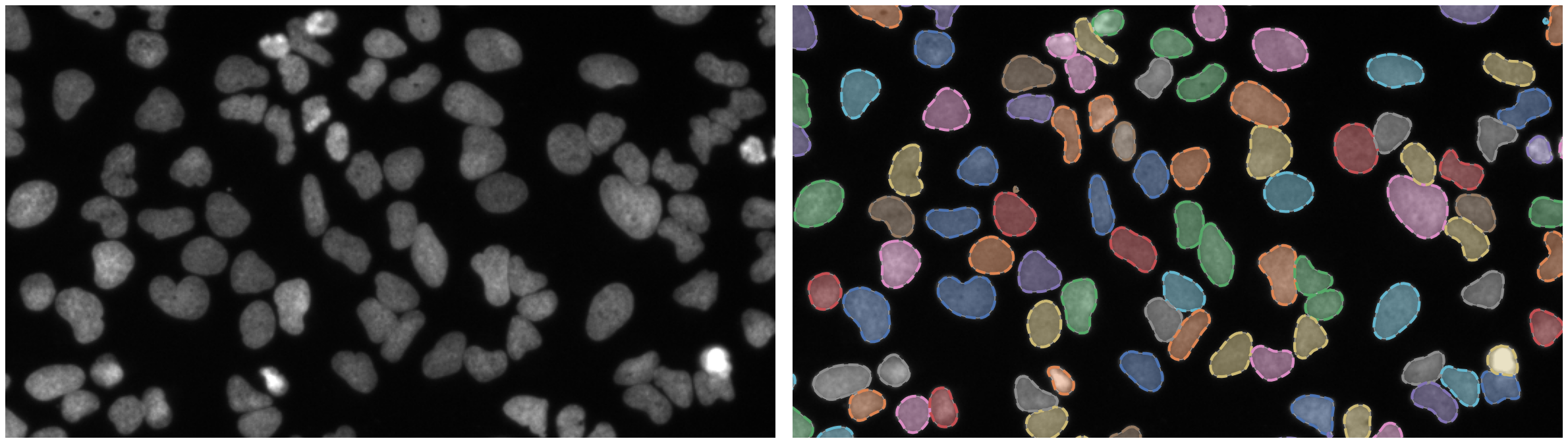
Nuclei of U2OS cells in a chemical screen
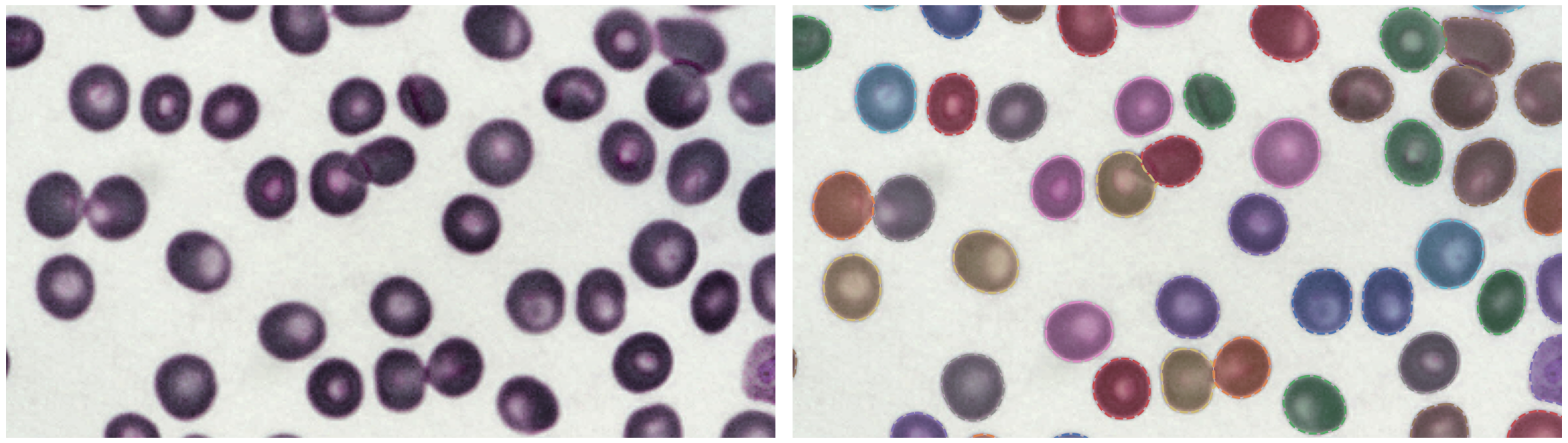
P. vivax (malaria) infected human blood
🛠 Install
Make sure you have PyTorch installed.
PyPI
pip install -U celldetection
GitHub
pip install git+https://github.com/FZJ-INM1-BDA/celldetection.git
💾 Trained models
model = cd.fetch_model(model_name, check_hash=True)
| model name | training data | link |
|---|---|---|
ginoro_CpnResNeXt101UNet-fbe875f1a3e5ce2c |
BBBC039, BBBC038, Omnipose, Cellpose, Sartorius - Cell Instance Segmentation, Livecell, NeurIPS 22 CellSeg Challenge | 🔗 |
Run a demo with a pretrained model
import torch, cv2, celldetection as cd
from skimage.data import coins
from matplotlib import pyplot as plt
# Load pretrained model
device = 'cuda' if torch.cuda.is_available() else 'cpu'
model = cd.fetch_model('ginoro_CpnResNeXt101UNet-fbe875f1a3e5ce2c', check_hash=True).to(device)
model.eval()
# Load input
img = coins()
img = cv2.cvtColor(img, cv2.COLOR_GRAY2RGB)
print(img.dtype, img.shape, (img.min(), img.max()))
# Run model
with torch.no_grad():
x = cd.to_tensor(img, transpose=True, device=device, dtype=torch.float32)
x = x / 255 # ensure 0..1 range
x = x[None] # add batch dimension: Tensor[3, h, w] -> Tensor[1, 3, h, w]
y = model(x)
# Show results for each batch item
contours = y['contours']
for n in range(len(x)):
cd.imshow_row(x[n], x[n], figsize=(16, 9), titles=('input', 'contours'))
cd.plot_contours(contours[n])
plt.show()
🔬 Architectures
import celldetection as cd
Contour Proposal Networks
cd.models.CPNcd.models.CpnU22cd.models.CPNCorecd.models.CpnResUNetcd.models.CpnSlimU22cd.models.CpnWideU22cd.models.CpnResNet18FPNcd.models.CpnResNet34FPNcd.models.CpnResNet50FPNcd.models.CpnResNeXt50FPNcd.models.CpnResNet101FPNcd.models.CpnResNet152FPNcd.models.CpnResNet18UNetcd.models.CpnResNet34UNetcd.models.CpnResNet50UNetcd.models.CpnResNeXt101FPNcd.models.CpnResNeXt152FPNcd.models.CpnResNeXt50UNetcd.models.CpnResNet101UNetcd.models.CpnResNet152UNetcd.models.CpnResNeXt101UNetcd.models.CpnResNeXt152UNetcd.models.CpnWideResNet50FPNcd.models.CpnWideResNet101FPNcd.models.CpnMobileNetV3LargeFPNcd.models.CpnMobileNetV3SmallFPN
PyTorch Image Models (timm)
Also have a look at Timm Documentation.
import timm
timm.list_models(filter='*') # explore available models
Segmentation Models PyTorch (smp)
import segmentation_models_pytorch as smp
smp.encoders.get_encoder_names() # explore available models
encoder = cd.models.SmpEncoder(encoder_name='mit_b5', pretrained='imagenet')
Find a list of Smp Encoders in the smp documentation.
U-Nets
# U-Nets are available in 2D and 3D
import celldetection as cd
model = cd.models.ResNeXt50UNet(in_channels=3, out_channels=1, nd=3)
cd.models.U22cd.models.U17cd.models.U12cd.models.UNetcd.models.WideU22cd.models.SlimU22cd.models.ResUNetcd.models.UNetEncodercd.models.ResNet50UNetcd.models.ResNet18UNetcd.models.ResNet34UNetcd.models.ResNet152UNetcd.models.ResNet101UNetcd.models.ResNeXt50UNetcd.models.ResNeXt152UNetcd.models.ResNeXt101UNetcd.models.WideResNet50UNetcd.models.WideResNet101UNetcd.models.MobileNetV3SmallUNetcd.models.MobileNetV3LargeUNet
MA-Nets
# Many MA-Nets are available in 2D and 3D
import celldetection as cd
encoder = cd.models.ConvNeXtSmall(in_channels=3, nd=3)
model = cd.models.MaNet(encoder, out_channels=1, nd=3)
Feature Pyramid Networks
cd.models.FPNcd.models.ResNet18FPNcd.models.ResNet34FPNcd.models.ResNet50FPNcd.models.ResNeXt50FPNcd.models.ResNet101FPNcd.models.ResNet152FPNcd.models.ResNeXt101FPNcd.models.ResNeXt152FPNcd.models.WideResNet50FPNcd.models.WideResNet101FPNcd.models.MobileNetV3LargeFPNcd.models.MobileNetV3SmallFPN
ConvNeXt Networks
# ConvNeXt Networks are available in 2D and 3D
import celldetection as cd
model = cd.models.ConvNeXtSmall(in_channels=3, nd=3)
Residual Networks
# Residual Networks are available in 2D and 3D
import celldetection as cd
model = cd.models.ResNet50(in_channels=3, nd=3)
Mobile Networks
🐳 Docker
Find us on Docker Hub: https://hub.docker.com/r/ericup/celldetection
You can pull the latest version of celldetection via:
docker pull ericup/celldetection:latest
CPN inference via Docker with GPU
docker run --rm \
-v $PWD/docker/outputs:/outputs/ \
-v $PWD/docker/inputs/:/inputs/ \
-v $PWD/docker/models/:/models/ \
--gpus="device=0" \
celldetection:latest /bin/bash -c \
"python cpn_inference.py --tile_size=1024 --stride=768 --precision=32-true"
CPN inference via Docker with CPU
docker run --rm \
-v $PWD/docker/outputs:/outputs/ \
-v $PWD/docker/inputs/:/inputs/ \
-v $PWD/docker/models/:/models/ \
celldetection:latest /bin/bash -c \
"python cpn_inference.py --tile_size=1024 --stride=768 --precision=32-true --accelerator=cpu"
Apptainer
You can also pull our Docker images for the use with Apptainer (formerly Singularity) with this command:
apptainer pull --dir . --disable-cache docker://ericup/celldetection:latest
🤗 Hugging Face Spaces
Find us on Hugging Face and upload your own images for segmentation: https://huggingface.co/spaces/ericup/celldetection
There's also an API (Python & JavaScript), allowing you to utilize community GPUs (currently Nvidia A100) remotely!
Hugging Face API
Python
from gradio_client import Client
# Define inputs (local filename or URL)
inputs = 'https://raw.githubusercontent.com/scikit-image/scikit-image/main/skimage/data/coins.png'
# Set up client
client = Client("ericup/celldetection")
# Predict
overlay_filename, img_filename, h5_filename, csv_filename = client.predict(
inputs, # str: Local filepath or URL of your input image
# Model name
'ginoro_CpnResNeXt101UNet-fbe875f1a3e5ce2c',
# Custom Score Threshold (numeric value between 0 and 1)
False, .9, # bool: Whether to use custom setting; float: Custom setting
# Custom NMS Threshold
False, .3142, # bool: Whether to use custom setting; float: Custom setting
# Custom Number of Sample Points
False, 128, # bool: Whether to use custom setting; int: Custom setting
# Overlapping objects
True, # bool: Whether to allow overlapping objects
# API name (keep as is)
api_name="/predict"
)
# Example usage: Code below only shows how to use the results
from matplotlib import pyplot as plt
import celldetection as cd
import pandas as pd
# Read results from local temporary files
img = imread(img_filename)
overlay = imread(overlay_filename) # random colors per instance; transparent overlap
properties = pd.read_csv(csv_filename)
contours, scores, label_image = cd.from_h5(h5_filename, 'contours', 'scores', 'labels')
# Optionally display overlay
cd.imshow_row(img, img, figsize=(16, 9))
cd.imshow(overlay)
plt.show()
# Optionally display contours with text
cd.imshow_row(img, img, figsize=(16, 9))
cd.plot_contours(contours, texts=['score: %d%%\narea: %d' % s for s in zip((scores * 100).round(), properties.area)])
plt.show()
Javascript
import { client } from "@gradio/client";
const response_0 = await fetch("https://raw.githubusercontent.com/scikit-image/scikit-image/main/skimage/data/coins.png");
const exampleImage = await response_0.blob();
const app = await client("ericup/celldetection");
const result = await app.predict("/predict", [
exampleImage, // blob: Your input image
// Model name (hosted model or URL)
"ginoro_CpnResNeXt101UNet-fbe875f1a3e5ce2c",
// Custom Score Threshold (numeric value between 0 and 1)
false, .9, // bool: Whether to use custom setting; float: Custom setting
// Custom NMS Threshold
false, .3142, // bool: Whether to use custom setting; float: Custom setting
// Custom Number of Sample Points
false, 128, // bool: Whether to use custom setting; int: Custom setting
// Overlapping objects
true, // bool: Whether to allow overlapping objects
// API name (keep as is)
api_name="/predict"
]);
🧑💻 Napari Plugin
Find our Napari Plugin here: https://github.com/FZJ-INM1-BDA/celldetection-napari
Find out more about Napari here: https://napari.org
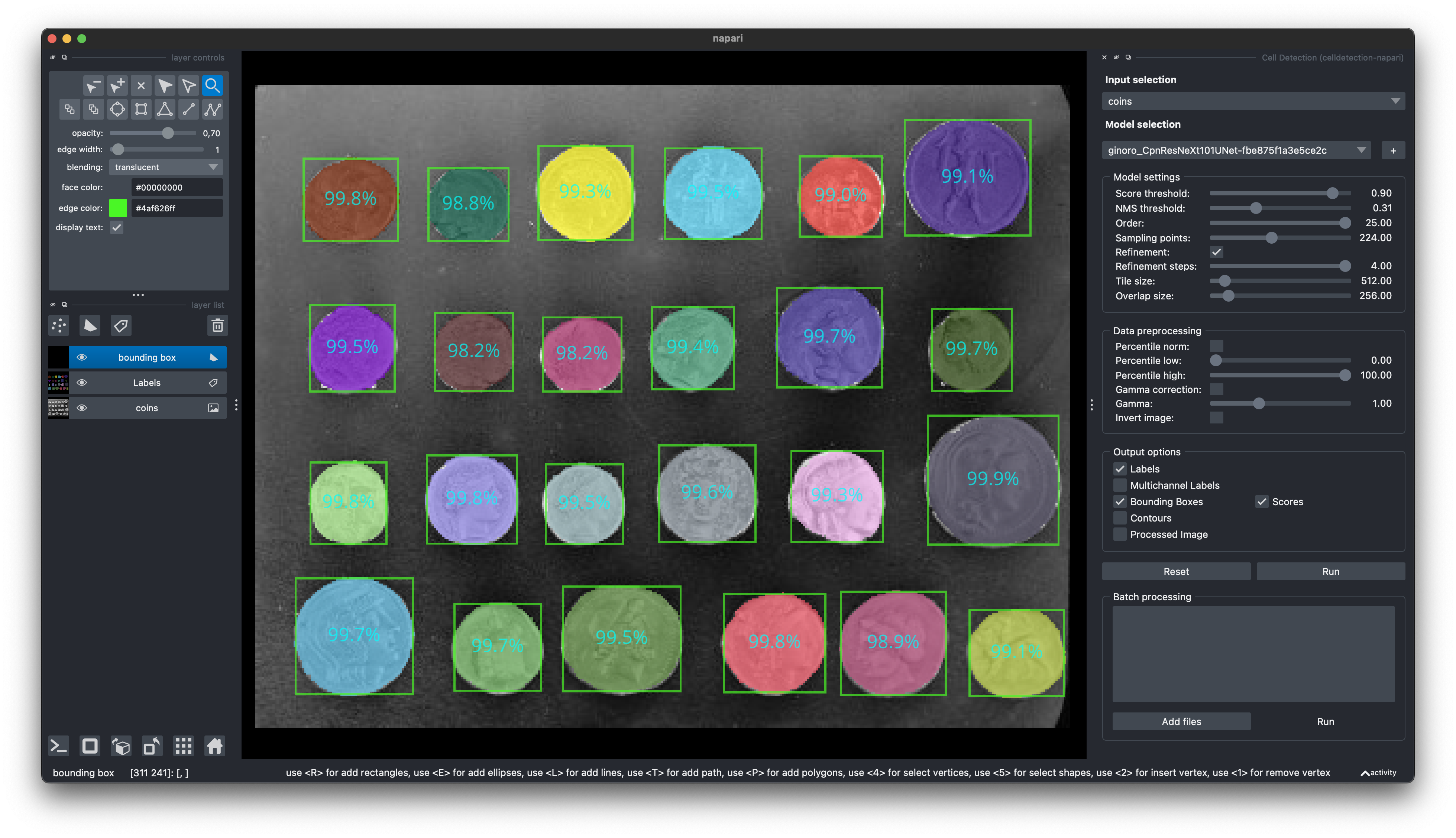
pip install git+https://github.com/FZJ-INM1-BDA/celldetection-napari.git
🏆 Awards
- NeurIPS 2022 Cell Segmentation Challenge: Winner Finalist Award
📝 Citing
If you find this work useful, please consider giving a star ⭐️ and citation:
@article{UPSCHULTE2022102371,
title = {Contour proposal networks for biomedical instance segmentation},
journal = {Medical Image Analysis},
volume = {77},
pages = {102371},
year = {2022},
issn = {1361-8415},
doi = {https://doi.org/10.1016/j.media.2022.102371},
url = {https://www.sciencedirect.com/science/article/pii/S136184152200024X},
author = {Eric Upschulte and Stefan Harmeling and Katrin Amunts and Timo Dickscheid},
keywords = {Cell detection, Cell segmentation, Object detection, CPN},
}
🔗 Links
🧑🔬 Thanks!
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
Filter files by name, interpreter, ABI, and platform.
If you're not sure about the file name format, learn more about wheel file names.
Copy a direct link to the current filters
File details
Details for the file celldetection-0.4.9.tar.gz.
File metadata
- Download URL: celldetection-0.4.9.tar.gz
- Upload date:
- Size: 153.7 kB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/5.1.0 CPython/3.9.19
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
71ec9876f927c32aea2754b4fdc4ee4554db734c40febe868deeafd095ee8654
|
|
| MD5 |
e769d9543526b68fbe6abb5711e5b5b7
|
|
| BLAKE2b-256 |
0ba4095705044694ad8a2bdf91489f0afed073dc50256b73843cce0328e6effc
|
File details
Details for the file CellDetection-0.4.9-py3-none-any.whl.
File metadata
- Download URL: CellDetection-0.4.9-py3-none-any.whl
- Upload date:
- Size: 172.5 kB
- Tags: Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/5.1.0 CPython/3.9.19
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
7151c61e545228079936c05fc92016df5d6c52cf6aab592ef734418d0f7d34d3
|
|
| MD5 |
7ea7e54c6f597a2700f28d6a93627e3f
|
|
| BLAKE2b-256 |
8c8c2fa078f3b87ca2b9317579f03b6feb1b6bad4231e0f7f7d133ec4524a5f5
|


















