Conserved water search is a tool for finding conserved water molecules from MD trajectories.
Project description
ConservedWaterSearch



The ConservedWaterSearch (CWS) Python library uses density based clustering approach to detect conserved waters from simulation trajectories. First, positions of water molecules are determined based on clustering of oxygen atoms belonging to water molecules(see figure below for more information). Positions on water molecules can be determined using Multi Stage Re-Clustering (MSRC) approach or Single Clustering (SC) approach (see for more information on clustering procedures).
Requirements: Python 3.10+ and NumPy 2+.
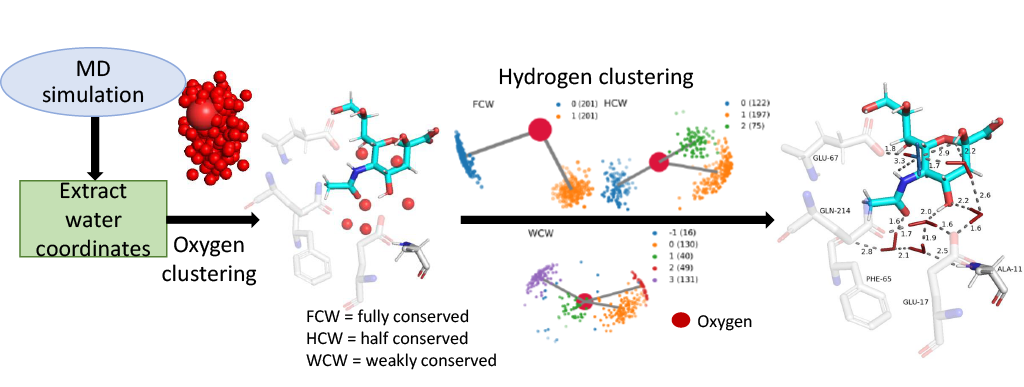
Conserved water molecules can be classified into 3 distinct conserved water types based on their hydrogen orientation: Fully Conserved Waters (FCW), Half Conserved Waters (HCW) and Weakly Conserved Waters (WCW) - see figure below for examples and more information or see CWS docs.
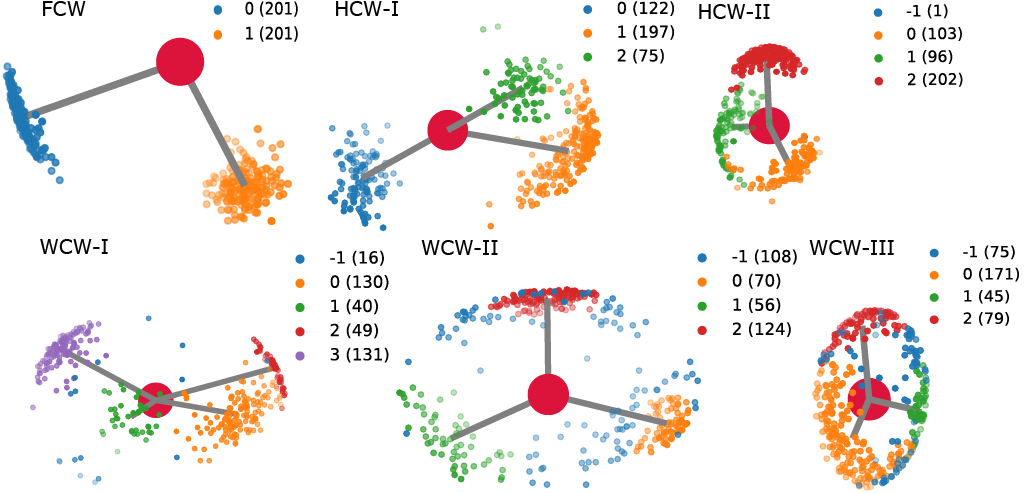
Both, MSRC and SC can be used with either OPTICS (via sklearn) and HDBSCAN. MSRC approach using either of the two algorithms produces better quality results at the cost of computational time, while SC approach produces lowe quality results at a fraction of the computational cost.
Important links
Documentation: hosted on Read The Docs
GitHub repository: source code/contribute code
Issue tracker: Report issues/request features
Citation
See this article.
Installation
The easiest ways to install ConservedWaterSearch is to install it from conda-forge using conda:
conda install -c conda-forge ConservedWaterSearchCWS can also be installed from PyPI (using pip). To install via pip use:
pip install ConservedWaterSearchOptional visualization dependencies
nglview can be installed from PyPI (pip) or conda or when installing CWS
through pip by using pip install ConservedWaterSearch[nglview].
PyMOL is the recomended visualization tool for CWS and can be installed
only using conda or from source. PyMOL is not available via PyPI
(pip), but can be installed from conda-forge. If PyMOL is
already installed in your current python environment it can be used
with CWS. If not, the free (open-source) version can be installed from
conda-forge via conda (or
mamba):
conda install -c conda-forge pymol-open-sourceand paid (licensed version) from schrodinger channel (see here for more details) via conda (or
mamba):
conda install -c conda-forge -c schrodinger pymol-bundleMatplotlib is only required for analyzing of clustering and is useful
if default values of clustering parameters need to be fine-tuned (which
should be relatively rarely). You can install it from pip or conda or
when installing CWS through pip by using pip install ConservedWaterSearch[matplotlib].
Both mpl and nglveiw can be installed when installing CWS by using:
pip install ConservedWaterSearch[all]For more information see installation.
Example
The easiest way to use CWS is by calling WaterClustering class. The starting trajectory should be aligned first, and coordinates of water oxygen and hydrogens extracted. See WaterNetworkAnalysis for more information and convenience functions.
# imports
from ConservedWaterSearch.water_clustering import WaterClustering
from ConservedWaterSearch.utils import get_orientations_from_positions
# Number of snapshots
Nsnap = 20
# load some example - trajectory should be aligned prior to extraction of atom coordinates
Opos = np.loadtxt("tests/data/testdataO.dat")
Hpos = np.loadtxt("tests/data/testdataH.dat")
wc = WaterClustering(nsnaps=Nsnap)
wc.multi_stage_reclustering(*get_orientations_from_positions(Opos, Hpos))
print(wc.water_type)
# "aligned.pdb" should be the snapshot original trajectory was aligned to.
wc.visualise_pymol(aligned_protein = "aligned.pdb", output_file = "waters.pse")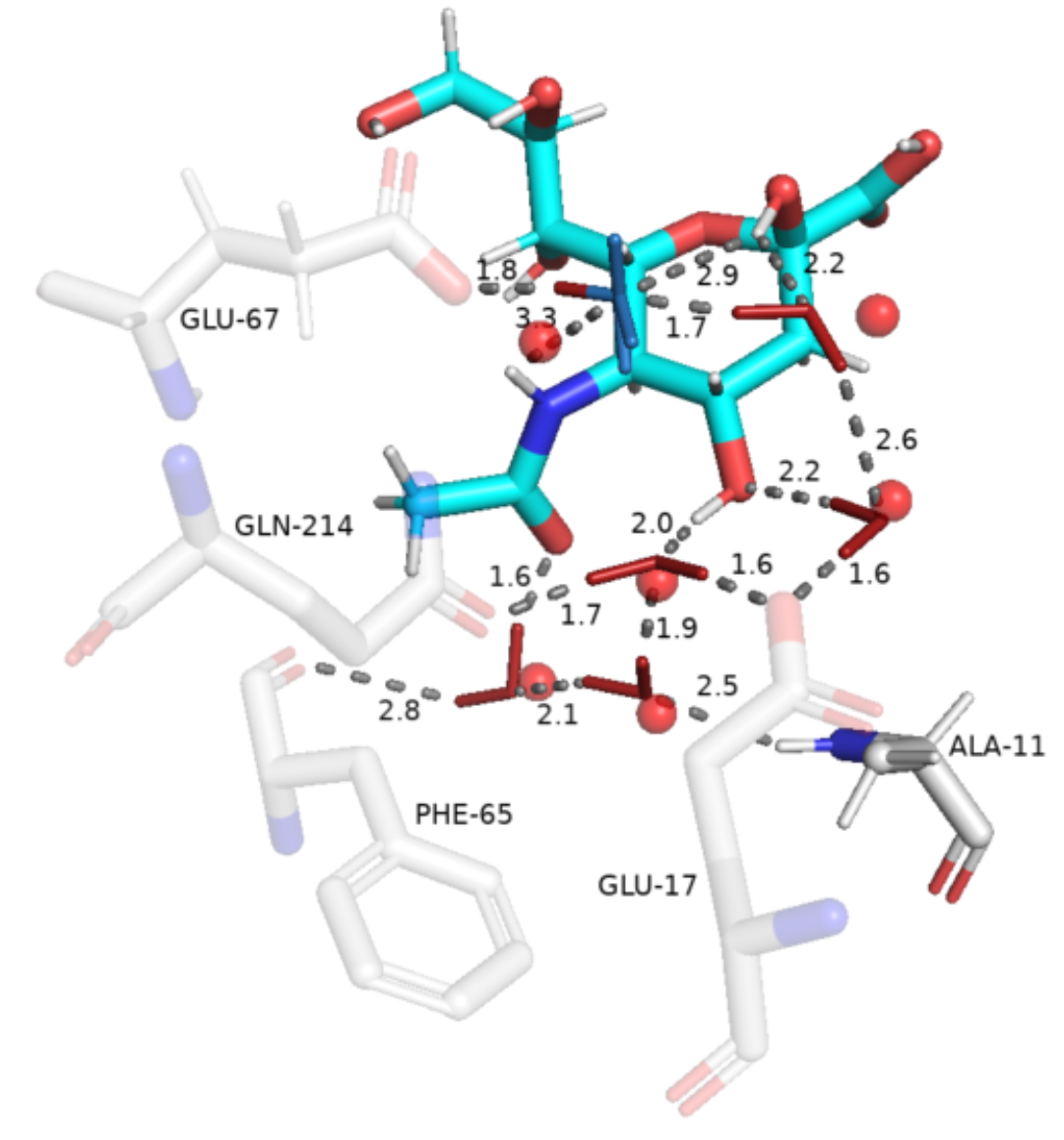
Sometimes users might want to explicitly classify conserved water molecules. A simple python code can be used to classify waters into categories given an array of 3D oxygen coordinates and their related relative hydrogen orientations:
import ConservedWaterSearch.hydrogen_orientation as HO
# load some example
orientations = np.loadtxt("tests/data/conserved_sample_FCW.dat")
# Run classification
res = HO.hydrogen_orientation_analysis(
orientations,
)
# print the water type
print(res[0][2])For more information on preprocessing trajectory data, please refer to the WaterNetworkAnalysis.
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
Filter files by name, interpreter, ABI, and platform.
If you're not sure about the file name format, learn more about wheel file names.
Copy a direct link to the current filters
File details
Details for the file conservedwatersearch-0.5.1.tar.gz.
File metadata
- Download URL: conservedwatersearch-0.5.1.tar.gz
- Upload date:
- Size: 33.9 kB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/6.1.0 CPython/3.13.7
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
b3601f370f85bbbc491a4d030cf8882a8dc23e76282d033b82be50ff9caa0bfe
|
|
| MD5 |
ce63740c2237ce0392cd9117f2454366
|
|
| BLAKE2b-256 |
e5db05e3753c532a562eca725bc462a021f6bdd8d662573bcb4f78e36838de89
|
File details
Details for the file conservedwatersearch-0.5.1-py3-none-any.whl.
File metadata
- Download URL: conservedwatersearch-0.5.1-py3-none-any.whl
- Upload date:
- Size: 30.8 kB
- Tags: Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/6.1.0 CPython/3.13.7
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
09ba569e95dbf2a1759a0a5687cff800e3d06c6304d2e86425e7832680df3768
|
|
| MD5 |
1e266a052ea6c63001e03650652a99ea
|
|
| BLAKE2b-256 |
b4d1ebe68f13ea1444025bd1b45854f1a59d8f06af3498702f4e00ebd4a0e33e
|











